

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137823.6 + phase: 0
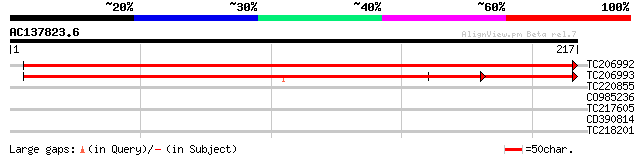
(217 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC206992 weakly similar to GB|BAB08647.1|9758068|AB009048 frnE p... 369 e-103
TC206993 similar to GB|BAB08647.1|9758068|AB009048 frnE protein-... 297 2e-81
TC220855 similar to GB|AAQ56831.1|34098897|BT010388 At3g03960 {A... 28 3.2
CO985236 27 7.1
TC217605 27 7.1
CD390814 similar to GP|13122434|dbj P0666G04.17 {Oryza sativa (j... 27 7.1
TC218201 similar to UP|Q948J9 (Q948J9) Lipoyltransferase, partia... 27 9.3
>TC206992 weakly similar to GB|BAB08647.1|9758068|AB009048 frnE protein-like
{Arabidopsis thaliana;} , partial (91%)
Length = 933
Score = 369 bits (947), Expect = e-103
Identities = 178/212 (83%), Positives = 197/212 (91%)
Frame = +3
Query: 6 SAHSEKKLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGI 65
S SEKKLVRID+SSDTVCPWCFVGKKNLDKAIA+SKDKYNFEI+WHP+QLSPDAPKEGI
Sbjct: 81 SGTSEKKLVRIDVSSDTVCPWCFVGKKNLDKAIAASKDKYNFEIIWHPFQLSPDAPKEGI 260
Query: 66 EKREFYRSKFGSRSDQMEARMSEVFRTVGLVYSLSGLTGNTMDSHRLIYFSGQQGLDKQH 125
+KRE+YR KFGS+S++MEARMSEVF+ VGL YSLSGLTGNT+DSHRLIYF+ QQGLDKQH
Sbjct: 261 DKREYYRRKFGSQSERMEARMSEVFKNVGLEYSLSGLTGNTIDSHRLIYFARQQGLDKQH 440
Query: 126 DLVEELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNGLKEVEEELKTYSRNI 185
DLVEEL +GYFTQ KYIGD +FLLE+AAKVGIEGAEEFLK+PNNGLKEVEEELK S NI
Sbjct: 441 DLVEELNIGYFTQGKYIGDHKFLLESAAKVGIEGAEEFLKDPNNGLKEVEEELKNNSGNI 620
Query: 186 TGVPYYVINGSQKLSGGQPPEVFLRAFQAATS 217
+GVPYYVING+ KLSGGQ PEVFLRAFQ AT+
Sbjct: 621 SGVPYYVINGTHKLSGGQAPEVFLRAFQVATT 716
>TC206993 similar to GB|BAB08647.1|9758068|AB009048 frnE protein-like
{Arabidopsis thaliana;} , partial (75%)
Length = 1033
Score = 297 bits (761), Expect = 2e-81
Identities = 152/219 (69%), Positives = 168/219 (76%), Gaps = 42/219 (19%)
Frame = +1
Query: 6 SAHSEKKLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGI 65
S SEKKL+RID S+DTVCPWCFVGKKNLDKAIASSKDKYNFEI+WHP+QL+PDAPKEGI
Sbjct: 109 SGISEKKLIRIDASADTVCPWCFVGKKNLDKAIASSKDKYNFEIIWHPFQLNPDAPKEGI 288
Query: 66 EKREFYRSKFGSRSDQMEARMSEVFRTVGLVYSLSGLT---------------------- 103
+KREFYRSKFGS+S++MEARMSEVF+ VGL YSLSGLT
Sbjct: 289 DKREFYRSKFGSQSERMEARMSEVFKNVGLEYSLSGLT*D*TWFYTSSMDKLI*TTLEEE 468
Query: 104 --------------------GNTMDSHRLIYFSGQQGLDKQHDLVEELGLGYFTQEKYIG 143
GNT+DSHRLIYF+ QQGLDKQHDLVEEL +GYFTQ KYIG
Sbjct: 469 CSEEEFSN*GSTTIVLIIHRGNTIDSHRLIYFARQQGLDKQHDLVEELNIGYFTQGKYIG 648
Query: 144 DQEFLLEAAAKVGIEGAEEFLKNPNNGLKEVEEELKTYS 182
D +FLLE+AAKVGIEGAEEFLK+PNNGLKEVEEELKTYS
Sbjct: 649 DHKFLLESAAKVGIEGAEEFLKDPNNGLKEVEEELKTYS 765
Score = 72.0 bits (175), Expect = 2e-13
Identities = 34/57 (59%), Positives = 41/57 (71%)
Frame = +2
Query: 161 EEFLKNPNNGLKEVEEELKTYSRNITGVPYYVINGSQKLSGGQPPEVFLRAFQAATS 217
+ FL+ P + + K RNI+GVPYYVING+ KLSGGQPPEVFLRAFQ AT+
Sbjct: 701 KSFLRTPTMV*RRLRRSSKLTQRNISGVPYYVINGNHKLSGGQPPEVFLRAFQVATN 871
>TC220855 similar to GB|AAQ56831.1|34098897|BT010388 At3g03960 {Arabidopsis
thaliana;} , partial (28%)
Length = 575
Score = 28.1 bits (61), Expect = 3.2
Identities = 20/79 (25%), Positives = 38/79 (47%)
Frame = +2
Query: 121 LDKQHDLVEELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNGLKEVEEELKT 180
L+ QH + L L Q++ IGD L + A ++GAEE ++ + + + K
Sbjct: 311 LEVQHPAAKVLVLAGKAQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKA 490
Query: 181 YSRNITGVPYYVINGSQKL 199
++ + + V NGS+ +
Sbjct: 491 INKTVQILDELVENGSESM 547
>CO985236
Length = 466
Score = 26.9 bits (58), Expect = 7.1
Identities = 17/65 (26%), Positives = 31/65 (47%), Gaps = 7/65 (10%)
Frame = -1
Query: 122 DKQHDLVEEL-------GLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNGLKEV 174
DK+ D VEE+ G+G ++K + + E ++G ++ + LKE+
Sbjct: 412 DKKEDGVEEVNEDRKEDGVGEVKEDKEVDGVSCVKEVEEDGEVDGVKKVEDKKGDELKEI 233
Query: 175 EEELK 179
EE+ K
Sbjct: 232 EEDKK 218
>TC217605
Length = 691
Score = 26.9 bits (58), Expect = 7.1
Identities = 12/34 (35%), Positives = 20/34 (58%)
Frame = -1
Query: 75 FGSRSDQMEARMSEVFRTVGLVYSLSGLTGNTMD 108
F S+S++++ V G++YSL TGN +D
Sbjct: 496 FYSKSNRLQKIFHPVIPPHGIIYSLDLFTGNNLD 395
>CD390814 similar to GP|13122434|dbj P0666G04.17 {Oryza sativa (japonica
cultivar-group)}, partial (14%)
Length = 492
Score = 26.9 bits (58), Expect = 7.1
Identities = 10/16 (62%), Positives = 11/16 (68%)
Frame = -2
Query: 46 NFEILWHPYQLSPDAP 61
NFE LW +Q PDAP
Sbjct: 104 NFEFLWTHFQFFPDAP 57
>TC218201 similar to UP|Q948J9 (Q948J9) Lipoyltransferase, partial (32%)
Length = 863
Score = 26.6 bits (57), Expect = 9.3
Identities = 10/27 (37%), Positives = 17/27 (62%), Gaps = 1/27 (3%)
Frame = -2
Query: 26 WCFVGKKNLDKAIASSKDKYNFEI-LW 51
W F+ KK +D+ + +KYNF + +W
Sbjct: 532 WPFIWKK*IDRGLLCISNKYNFAVSIW 452
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,390,451
Number of Sequences: 63676
Number of extensions: 96457
Number of successful extensions: 313
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 312
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 313
length of query: 217
length of database: 12,639,632
effective HSP length: 93
effective length of query: 124
effective length of database: 6,717,764
effective search space: 833002736
effective search space used: 833002736
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 57 (26.6 bits)
Medicago: description of AC137823.6


