

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137823.1 + phase: 0
(249 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
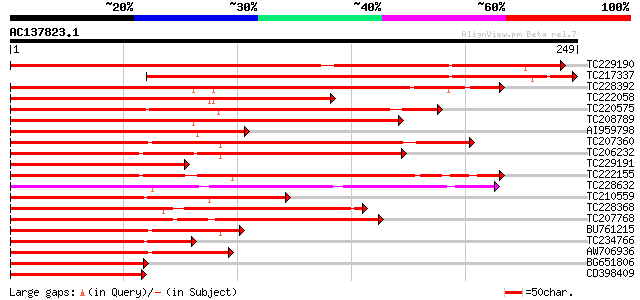
Score E
Sequences producing significant alignments: (bits) Value
TC229190 similar to UP|Q9SEG4 (Q9SEG4) CAGL2, partial (86%) 385 e-107
TC217337 similar to UP|Q9ARF0 (Q9ARF0) MADS2 protein (Fragment),... 310 5e-85
TC228392 similar to UP|Q8LLR0 (Q8LLR0) MADS-box protein 4, parti... 273 7e-74
TC222058 homologue to UP|AGL9_PETHY (Q03489) Agamous-like MADS b... 230 5e-61
TC220575 similar to UP|Q8LLR1 (Q8LLR1) MADS-box protein 3, parti... 171 4e-43
TC208789 similar to UP|Q9SBQ1 (Q9SBQ1) MADS box transcription fa... 167 5e-42
AI959798 similar to GP|22091481|emb MADS box transcription facto... 160 7e-40
TC207360 similar to UP|Q8VWZ3 (Q8VWZ3) C-type MADS box protein, ... 158 2e-39
TC206232 homologue to UP|Q8H6F8 (Q8H6F8) MADS box protein GHMADS... 155 2e-38
TC229191 homologue to UP|Q84U54 (Q84U54) MADS-RIN-like protein, ... 152 2e-37
TC222155 weakly similar to UP|O81662 (O81662) Transcription acti... 142 1e-34
TC228632 similar to UP|Q9ZTQ9 (Q9ZTQ9) MADS box protein 26, part... 125 2e-29
TC210559 similar to UP|Q84MI1 (Q84MI1) MADS-box protein (Fragmen... 118 2e-27
TC228368 similar to UP|O49173 (O49173) MADS-box protein NMH 7, p... 115 2e-26
TC207768 similar to UP|Q9ST53 (Q9ST53) MADS-box protein 4, parti... 114 4e-26
BU761215 homologue to PIR|T08039|T08 MADS-box protein - cucumber... 112 2e-25
TC234766 homologue to UP|Q7Y137 (Q7Y137) MADS-box protein PTM5, ... 104 4e-23
AW706936 99 2e-21
BG651806 92 3e-19
CD398409 91 5e-19
>TC229190 similar to UP|Q9SEG4 (Q9SEG4) CAGL2, partial (86%)
Length = 1062
Score = 385 bits (988), Expect = e-107
Identities = 202/248 (81%), Positives = 216/248 (86%), Gaps = 4/248 (1%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST
Sbjct: 313 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 492
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKELESSYREYLKLKQRFENLQRAQRNLLGEDLGPLS 120
++MLKTL+RYQKCSYGAVEVSKP KELESSYREYLKLK RFE+LQR QRNLLGEDLGPL+
Sbjct: 493 NSMLKTLERYQKCSYGAVEVSKPGKELESSYREYLKLKARFESLQRTQRNLLGEDLGPLN 672
Query: 121 SKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEEINSRNHY 180
+KDLEQLERQLDSSLKQ TQFMLDQLADLQNKEHMLVEANRSL++KLEEINSRN Y
Sbjct: 673 TKDLEQLERQLDSSLKQ-----TQFMLDQLADLQNKEHMLVEANRSLTMKLEEINSRNQY 837
Query: 181 RQSWEASDQSMQYEAQQNAHSQSFFQQLECNPTLQIGSDYRYNNV----ASDQIASTSQA 236
RQ+WEA +QSM Y QNAHSQ FFQ LECNPTLQIGSDYRY + S +A+T
Sbjct: 838 RQTWEAGEQSMSY-GTQNAHSQGFFQPLECNPTLQIGSDYRYQPLKALRTSS*LATTXSF 1014
Query: 237 QQQVNGFV 244
Q GF+
Sbjct: 1015NQVKWGFI 1038
>TC217337 similar to UP|Q9ARF0 (Q9ARF0) MADS2 protein (Fragment), partial
(88%)
Length = 980
Score = 310 bits (793), Expect = 5e-85
Identities = 159/190 (83%), Positives = 174/190 (90%), Gaps = 1/190 (0%)
Frame = +2
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKELESSYREYLKLKQRFENLQRAQRNLLGEDLGPLS 120
++MLKTL+RYQKCSYGAVEVSKP KELESSYREYLKLK RFE+LQR QRNLLGEDLGPL+
Sbjct: 2 NSMLKTLERYQKCSYGAVEVSKPGKELESSYREYLKLKARFESLQRTQRNLLGEDLGPLN 181
Query: 121 SKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEEINSRNHY 180
+KDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSL++KLEEINSRN Y
Sbjct: 182 TKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLTMKLEEINSRNQY 361
Query: 181 RQSWEASDQSMQYEAQQNAHSQSFFQQLECNPTLQIGSDYRYNNVASD-QIASTSQAQQQ 239
RQ+WEA +QSM Y QNAHSQ FFQ LECNPTLQIGSDYRY AS+ Q+A+T+QA QQ
Sbjct: 362 RQTWEAGEQSMPY-GTQNAHSQGFFQPLECNPTLQIGSDYRYIPEASEQQLAATTQA-QQ 535
Query: 240 VNGFVPGWML 249
VNGF+PGWML
Sbjct: 536 VNGFIPGWML 565
>TC228392 similar to UP|Q8LLR0 (Q8LLR0) MADS-box protein 4, partial (98%)
Length = 755
Score = 273 bits (697), Expect = 7e-74
Identities = 152/222 (68%), Positives = 174/222 (77%), Gaps = 5/222 (2%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFS RGK YEFCS
Sbjct: 40 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKQYEFCSG 219
Query: 61 SNMLKTLDRYQKCSYGAVE---VSKPAKELE-SSYREYLKLKQRFENLQRAQRNLLGEDL 116
S+MLKTL+RYQKC+YGA E +K A LE SS +EYL+LK R+E LQR+QRNL+GEDL
Sbjct: 220 SSMLKTLERYQKCNYGAPEDNVATKEALVLELSSQQEYLRLKARYEALQRSQRNLMGEDL 399
Query: 117 GPLSSKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEEINS 176
GPLSSK+LE LERQLDSSLKQ+RS +TQFMLDQL+DLQ KEH L E+NR L +LEE
Sbjct: 400 GPLSSKELESLERQLDSSLKQIRSIRTQFMLDQLSDLQRKEHFLGESNRDLRQRLEEFQI 579
Query: 177 RNHYRQSWEASDQSM-QYEAQQNAHSQSFFQQLECNPTLQIG 217
N + + A + ++ Q H + FQ +EC P LQIG
Sbjct: 580 -NPLQLNPSAEEVGYGRHPXQPQGH--AVFQPVECEPALQIG 696
>TC222058 homologue to UP|AGL9_PETHY (Q03489) Agamous-like MADS box protein
AGL9 homolog (Floral homeotic protein FBP2) (Floral
binding protein 2), partial (60%)
Length = 495
Score = 230 bits (586), Expect = 5e-61
Identities = 120/145 (82%), Positives = 133/145 (90%), Gaps = 2/145 (1%)
Frame = +3
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFS RGKLYEFCS+
Sbjct: 60 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 239
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKE-LE-SSYREYLKLKQRFENLQRAQRNLLGEDLGP 118
S+MLKTL+RYQKC+YGA E + +E LE SS +EYLKLK R+E LQR+QRNL+GEDLGP
Sbjct: 240 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 419
Query: 119 LSSKDLEQLERQLDSSLKQVRSTKT 143
LSSK+LE LERQLDSSLKQ+RST+T
Sbjct: 420 LSSKELESLERQLDSSLKQIRSTRT 494
>TC220575 similar to UP|Q8LLR1 (Q8LLR1) MADS-box protein 3, partial (44%)
Length = 851
Score = 171 bits (432), Expect = 4e-43
Identities = 93/192 (48%), Positives = 131/192 (67%), Gaps = 2/192 (1%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRG+V L+RI+NKINRQVTF+KRRNGLLKKA+ELSVLCDAE+AL+IFS+RGKL+++ ST
Sbjct: 158 MGRGKVVLERIQNKINRQVTFSKRRNGLLKKAFELSVLCDAEIALVIFSSRGKLFQYSST 337
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKELESS--YREYLKLKQRFENLQRAQRNLLGEDLGP 118
++ + +++Y++C + + A+ Y+E L L+ + E+LQR QRNLLGE+L P
Sbjct: 338 -DINRIIEKYRQCCFNMSQTGDVAEHQSEQCLYQELLILRVKHESLQRTQRNLLGEELEP 514
Query: 119 LSSKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEEINSRN 178
LS K+L LE+QLD +L Q R TQ ++ ++ +L K H L + N+ L E R
Sbjct: 515 LSMKELHSLEKQLDRTLGQARKHLTQKLISRIDELHGKVHNLEQVNKHL-----ESQERG 679
Query: 179 HYRQSWEASDQS 190
Q+ E S S
Sbjct: 680 KCTQTCEGSANS 715
>TC208789 similar to UP|Q9SBQ1 (Q9SBQ1) MADS box transcription factor,
partial (72%)
Length = 995
Score = 167 bits (422), Expect = 5e-42
Identities = 93/174 (53%), Positives = 123/174 (70%), Gaps = 1/174 (0%)
Frame = +3
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRGRVELKRIENKI QVTF+KRR+GLLKKA E+SVLCDA+VALI+FST+GKL ++ +
Sbjct: 219 MGRGRVELKRIENKIFMQVTFSKRRSGLLKKAREISVLCDADVALIVFSTKGKLLDYSNQ 398
Query: 61 SNMLKTLDRYQKCSYGAVE-VSKPAKELESSYREYLKLKQRFENLQRAQRNLLGEDLGPL 119
+ L+RY++ SY + V E+ E+ KLK R E LQR QRN +GEDL L
Sbjct: 399 PCTERILERYERYSYAERQLVGDDQPPNENWVIEHEKLKARVEVLQRNQRNFMGEDLDSL 578
Query: 120 SSKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEE 173
+ + L+ LE+QLDS+LK +RS K Q M + +++LQ K+ L E N LS K++E
Sbjct: 579 NLRGLQSLEQQLDSALKHIRSRKNQAMNESISELQKKDRTLREHNNLLSKKIKE 740
>AI959798 similar to GP|22091481|emb MADS box transcription factor {Daucus
carota subsp. sativus}, partial (39%)
Length = 409
Score = 160 bits (404), Expect = 7e-40
Identities = 82/106 (77%), Positives = 90/106 (84%), Gaps = 1/106 (0%)
Frame = +3
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRG+VELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFS RGKLYEFCS
Sbjct: 90 MGRGKVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSG 269
Query: 61 SNMLKTLDRYQKCSYGAVEVS-KPAKELESSYREYLKLKQRFENLQ 105
+ KTL+RY +CSYGA+EV +P E + Y+EYLKLK R E LQ
Sbjct: 270 HSTAKTLERYHRCSYGALEVQHQPEIETQRRYQEYLKLKSRVEALQ 407
>TC207360 similar to UP|Q8VWZ3 (Q8VWZ3) C-type MADS box protein, partial
(88%)
Length = 1075
Score = 158 bits (400), Expect = 2e-39
Identities = 90/205 (43%), Positives = 130/205 (62%), Gaps = 1/205 (0%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRG++E+KRIEN NRQVTF KRRNGLLKKAYELSVLCDAEVAL++FSTRG+LYE+ +
Sbjct: 95 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVVFSTRGRLYEYANN 274
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKELESSY-REYLKLKQRFENLQRAQRNLLGEDLGPL 119
S + T++RY+K + A ++ Y +E KL+++ ++Q R++LGE LG L
Sbjct: 275 S-VRATIERYKKANAAASNAESVSEANTQFYQQESSKLRRQIRDIQNLNRHILGEALGSL 451
Query: 120 SSKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEEINSRNH 179
S K+L+ LE +L+ L +VRS K + + + +Q +E L N L K+ E H
Sbjct: 452 SLKELKNLEGRLEKGLSRVRSRKHETLFADVEFMQKREIELQNHNNYLRAKIAE-----H 616
Query: 180 YRQSWEASDQSMQYEAQQNAHSQSF 204
R + S+ +M ++ SQS+
Sbjct: 617 ERAQQQQSNMNMSGTLCESLPSQSY 691
>TC206232 homologue to UP|Q8H6F8 (Q8H6F8) MADS box protein GHMADS-2, partial
(95%)
Length = 960
Score = 155 bits (392), Expect = 2e-38
Identities = 83/176 (47%), Positives = 121/176 (68%), Gaps = 2/176 (1%)
Frame = +3
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRG++E+KRIEN NRQVTF KRRNGLLKKAYELSVLCDAEVALI+FS+RG+LYE+ S
Sbjct: 75 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEY-SN 251
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKELESSY--REYLKLKQRFENLQRAQRNLLGEDLGP 118
+N+ T++RY+K S E+ + Y +E KL+Q+ + LQ + R+L+G+ L
Sbjct: 252 NNIRSTIERYKKACSDHSSAS-TTTEINAQYYQQESAKLRQQIQMLQNSNRHLMGDALST 428
Query: 119 LSSKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEEI 174
L+ K+L+QLE +L+ + ++RS K + +L ++ Q +E L N L K+ ++
Sbjct: 429 LTVKELKQLENRLERGITRIRSKKHEMLLAEIEYFQKREIELENENLCLRTKITDV 596
>TC229191 homologue to UP|Q84U54 (Q84U54) MADS-RIN-like protein, partial
(32%)
Length = 558
Score = 152 bits (383), Expect = 2e-37
Identities = 75/79 (94%), Positives = 78/79 (97%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFS RGKLYEFCS+
Sbjct: 320 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 499
Query: 61 SNMLKTLDRYQKCSYGAVE 79
S+MLKTL+RYQKCSYGAVE
Sbjct: 500 SSMLKTLERYQKCSYGAVE 556
>TC222155 weakly similar to UP|O81662 (O81662) Transcription activator,
partial (65%)
Length = 915
Score = 142 bits (359), Expect = 1e-34
Identities = 90/222 (40%), Positives = 135/222 (60%), Gaps = 5/222 (2%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
M RG+V+LK+IE+ +RQV F+KRR+GLLKKAYELSVLCDAEVA+I+FS G+LYEF S+
Sbjct: 152 MARGKVQLKKIEDTTSRQVAFSKRRSGLLKKAYELSVLCDAEVAVIVFSQNGRLYEF-SS 328
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKELESSYREYLK-----LKQRFENLQRAQRNLLGED 115
S+M K L+RY++ + PA + Y + LK L ++ E L+ ++R LLG+
Sbjct: 329 SDMTKILERYREHTKDV-----PASKFGDDYIQQLKLDSASLAKKIELLEHSKRELLGQS 493
Query: 116 LGPLSSKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEEIN 175
+ S +L+ +E QL SL++VR KTQ +Q+ L+++E L++ N LS +
Sbjct: 494 VSSCSYDELKGIEEQLQISLQRVRQRKTQLYTEQIDQLRSQESNLLKENAKLSAMWQRAE 673
Query: 176 SRNHYRQSWEASDQSMQYEAQQNAHSQSFFQQLECNPTLQIG 217
+ +Q W Q+ EA+ + S Q L+ + L IG
Sbjct: 674 KSS--QQQWPRHTQA---EAEPHCSSS---QSLDVDTELFIG 775
>TC228632 similar to UP|Q9ZTQ9 (Q9ZTQ9) MADS box protein 26, partial (82%)
Length = 880
Score = 125 bits (313), Expect = 2e-29
Identities = 79/216 (36%), Positives = 127/216 (58%), Gaps = 1/216 (0%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRG++E+KRIEN NRQVT++KR+NG+LKKA E++VLCDA+V+LIIF+ GK++++ S
Sbjct: 55 MGRGKIEIKRIENSSNRQVTYSKRKNGILKKAKEITVLCDAQVSLIIFAASGKMHDYISP 234
Query: 61 S-NMLKTLDRYQKCSYGAVEVSKPAKELESSYREYLKLKQRFENLQRAQRNLLGEDLGPL 119
S ++ L+RY K S + +K E+ E +LK+ +++Q R+L G+D+ L
Sbjct: 235 STTLIDILERYHKTSGKRLWDAKH----ENLNGEIERLKKENDSMQIELRHLKGDDINSL 402
Query: 120 SSKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEEINSRNH 179
+ K+L LE L++ L VR + +D L+ + +L E NR L+ ++ +
Sbjct: 403 NYKELMALEDALETGLVSVREKQ----MDVYRMLRRNDKILEEENRELNFLWQQRLAEEG 570
Query: 180 YRQSWEASDQSMQYEAQQNAHSQSFFQQLECNPTLQ 215
R+ DQS++ N+H F+ P LQ
Sbjct: 571 AREVDNGFDQSVR---DYNSHMPFAFRVQPMQPNLQ 669
>TC210559 similar to UP|Q84MI1 (Q84MI1) MADS-box protein (Fragment), partial
(64%)
Length = 710
Score = 118 bits (296), Expect = 2e-27
Identities = 65/124 (52%), Positives = 91/124 (72%), Gaps = 1/124 (0%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
M RG +++RIEN +RQVTF+KRRNGLLKKA+ELSVLCDAEVALIIFS RGKLYEF S
Sbjct: 340 MVRGETQMRRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALIIFSPRGKLYEFAS- 516
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKE-LESSYREYLKLKQRFENLQRAQRNLLGEDLGPL 119
S+M T++RY++ + A V++ ++ ++ +E L ++ E L+ ++R LLGE LG
Sbjct: 517 SSMQDTIERYRRHNRSAQTVNRSDEQNMQHLKQETANLMKKIELLEASKRKLLGEGLGSC 696
Query: 120 SSKD 123
S ++
Sbjct: 697 SLEE 708
>TC228368 similar to UP|O49173 (O49173) MADS-box protein NMH 7, partial (97%)
Length = 1042
Score = 115 bits (287), Expect = 2e-26
Identities = 66/158 (41%), Positives = 99/158 (61%), Gaps = 1/158 (0%)
Frame = +3
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
M RG++++KRIEN NRQVT++KRRNGL KKA EL+VLCDA+V++I+FS+ GKL+++ S
Sbjct: 81 MARGKIQIKRIENNTNRQVTYSKRRNGLFKKANELTVLCDAKVSIIMFSSTGKLHQYISP 260
Query: 61 SNMLKT-LDRYQKCSYGAVEVSKPAKELESSYREYLKLKQRFENLQRAQRNLLGEDLGPL 119
S K D+YQ + V E+ KLK+ NL++ R +G+ L L
Sbjct: 261 STSTKQFFDQYQM----TLGVDLWNSHYENMQENLKKLKEVNRNLRKEIRQRMGDCLNEL 428
Query: 120 SSKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKE 157
+DL+ LE ++D + K VR K + + +Q+ D Q K+
Sbjct: 429 GMEDLKLLEEEMDKAAKVVRERKYKVITNQI-DTQRKK 539
>TC207768 similar to UP|Q9ST53 (Q9ST53) MADS-box protein 4, partial (27%)
Length = 951
Score = 114 bits (285), Expect = 4e-26
Identities = 66/164 (40%), Positives = 101/164 (61%)
Frame = +3
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MG+ +VE+KRIENK RQ+TF+KRRNGL+KKA ELS+LCDA+VAL+IFS+ GKLYE C+
Sbjct: 111 MGKKKVEIKRIENKSTRQITFSKRRNGLMKKARELSILCDAKVALLIFSSTGKLYELCNG 290
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKELESSYREYLKLKQRFENLQRAQRNLLGEDLGPLS 120
++ + + +Y GA ++EL E + Q +R+ +L LS
Sbjct: 291 DSLAEVVQQYWD-HLGASGTDTKSQEL---CFEIADIWSGSAFSQMIKRHFGVSELEHLS 458
Query: 121 SKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEAN 164
DL +LE+ ++L ++RS K + M++ + +L+ K L + N
Sbjct: 459 VSDLMELEKLTHAALSRIRSAKMRLMMESVVNLKKKIEALEKTN 590
>BU761215 homologue to PIR|T08039|T08 MADS-box protein - cucumber, partial
(42%)
Length = 450
Score = 112 bits (279), Expect = 2e-25
Identities = 59/104 (56%), Positives = 77/104 (73%), Gaps = 1/104 (0%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRG++E+KRIEN NRQVTF KRRNGLLKKAYELSVLCDAEVALI+FS+RG+LYE+ +
Sbjct: 142 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANN 321
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKELESSY-REYLKLKQRFEN 103
S + T++RY+K S + ++ Y +E KL+Q+ N
Sbjct: 322 S-VKATIERYKKASSDSSGAGSASEANAQFYQQEADKLRQQISN 450
>TC234766 homologue to UP|Q7Y137 (Q7Y137) MADS-box protein PTM5, partial
(33%)
Length = 490
Score = 104 bits (259), Expect = 4e-23
Identities = 54/82 (65%), Positives = 68/82 (82%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
M RG+ +L+RIEN +RQVTF+KRRNGLLKKA+ELSVLCDAEVALIIFS RGKLYEF S
Sbjct: 239 MVRGKTQLRRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALIIFSPRGKLYEFAS- 415
Query: 61 SNMLKTLDRYQKCSYGAVEVSK 82
S+M T++RY++ + A V++
Sbjct: 416 SSMQDTIERYRRHNRSAQTVNR 481
>AW706936
Length = 422
Score = 99.0 bits (245), Expect = 2e-21
Identities = 50/98 (51%), Positives = 68/98 (69%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRG++ ++RI+N +RQVTF+KRRNGLLKKA ELS+LCDAEV L++FS+ GKLY++ ST
Sbjct: 131 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKARELSILCDAEVGLMVFSSTGKLYDYAST 310
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKELESSYREYLKLK 98
S M ++RY K + PA E + E L+
Sbjct: 311 S-MKSVIERYNKLKEEHHHLMNPASEAKFWQTEAASLR 421
>BG651806
Length = 309
Score = 91.7 bits (226), Expect = 3e-19
Identities = 41/61 (67%), Positives = 54/61 (88%)
Frame = +3
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRG++ ++RI+N +RQVTF+KRRNGLLKKA ELS+LCDAEV L++FS+ GKLY++ ST
Sbjct: 120 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKARELSILCDAEVGLMVFSSTGKLYDYAST 299
Query: 61 S 61
S
Sbjct: 300 S 302
>CD398409
Length = 639
Score = 90.9 bits (224), Expect = 5e-19
Identities = 41/60 (68%), Positives = 51/60 (84%)
Frame = -1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRG++ ++RIEN NRQVTF KRRNGLLKK ELS+LCDAEV +I+FS+ GKLYE+ +T
Sbjct: 549 MGRGKIPIRRIENSTNRQVTFCKRRNGLLKKTRELSILCDAEVGVIVFSSTGKLYEYSNT 370
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.129 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,654,583
Number of Sequences: 63676
Number of extensions: 94171
Number of successful extensions: 534
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 518
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 519
length of query: 249
length of database: 12,639,632
effective HSP length: 95
effective length of query: 154
effective length of database: 6,590,412
effective search space: 1014923448
effective search space used: 1014923448
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC137823.1


