

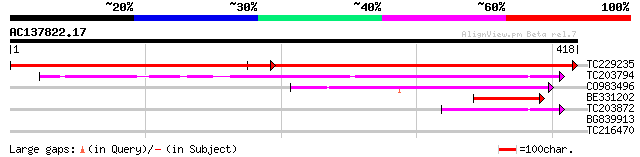
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137822.17 - phase: 0
(418 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC229235 similar to UP|Q9LHR6 (Q9LHR6) Arabidopsis thaliana geno... 400 e-112
TC203794 weakly similar to UP|Q8YV67 (Q8YV67) Alr2114 protein, p... 186 1e-47
CO983496 98 8e-21
BE331202 85 7e-17
TC203872 weakly similar to UP|Q6NVM1 (Q6NVM1) MGC69430 protein, ... 43 3e-04
BG839913 similar to GP|9955560|emb| protein kinase-like {Arabido... 29 4.5
TC216470 weakly similar to UP|Q9M6T9 (Q9M6T9) Lipid-transfer pro... 28 7.7
>TC229235 similar to UP|Q9LHR6 (Q9LHR6) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K3D20, partial (78%)
Length = 1379
Score = 400 bits (1029), Expect = e-112
Identities = 201/243 (82%), Positives = 215/243 (87%)
Frame = +2
Query: 176 GVLLFHEVGHFLAAFPKQVKLSIPFFIPHITLGSFGAITQFKSILPDRSTQVDISLAGPF 235
G F + F F +VKLSIP FIP+ITLGSFGAITQFKSILPDRSTQVDISLAGPF
Sbjct: 590 GFFCFMRLDTFYQHFLNKVKLSIPVFIPNITLGSFGAITQFKSILPDRSTQVDISLAGPF 769
Query: 236 AGAVLSFSMFAVGLLLSSNPDVAGDLVQVPSMLFQGSLLLGLISRATLGYAAVHAATVPI 295
AGAVLSFSMFAVGLLLSSNPD+ GDLVQVPS+LFQGSLLLGLISRATLGYAA+HA TVPI
Sbjct: 770 AGAVLSFSMFAVGLLLSSNPDITGDLVQVPSLLFQGSLLLGLISRATLGYAAMHAETVPI 949
Query: 296 HPLVIAGWCGLTIQAFNMLPVGCLDGGRSVQGAFGKGATMVFGLTTYTLLGLGVLGGPLS 355
HPLVIAGWCGLTIQAFNMLPVGCLDGGR+VQGAFGK A + FGLTTYTLLGLGVLGGPLS
Sbjct: 950 HPLVIAGWCGLTIQAFNMLPVGCLDGGRAVQGAFGKNALVGFGLTTYTLLGLGVLGGPLS 1129
Query: 356 LAWGFFVIFSQRSPEKPCLNDVTEVGTWRQTFVGVAIFLAVLTLLPVWDELAEELGIGLV 415
L WG +V+ QR+PEKPCLNDVTEVGTWR+ V +AIFL VLTL+PV DELAEELGIGLV
Sbjct: 1130LPWGLYVLLCQRAPEKPCLNDVTEVGTWRKALVAIAIFLVVLTLVPVGDELAEELGIGLV 1309
Query: 416 TTF 418
T F
Sbjct: 1310TAF 1318
Score = 342 bits (878), Expect = 1e-94
Identities = 165/196 (84%), Positives = 183/196 (93%)
Frame = +1
Query: 1 MELLGPEKVDPADVKKIKDKLFGYSTFWVTKEEPFGELGEGILFIGNLRGKREDIFSILQ 60
+ELLGPEKVDPADVK IKDKLFGYSTFWVTKEEPFG+LGEGILFIGNLRGKRED+F+ LQ
Sbjct: 64 IELLGPEKVDPADVKLIKDKLFGYSTFWVTKEEPFGDLGEGILFIGNLRGKREDVFAKLQ 243
Query: 61 NRLVEATGDKYNLFMVEEPDSDSPDPRGGPRVSFGLLRKEVSEPEETTLWQYVVASLLFL 120
N+LVE TGDKYNLFM EEP++DSPDPR GPRVSFGLLRKEVSEP TLWQYV+A LLFL
Sbjct: 244 NQLVEVTGDKYNLFMAEEPNADSPDPRSGPRVSFGLLRKEVSEPGPMTLWQYVIALLLFL 423
Query: 121 LTIGTSVEVGIASQINRLPPELVKFLTDPNYTEAPDMEMLYPFVESALPLAYGVLGVLLF 180
LTIG+SVE+GIASQINRLPPE+VK+ TDP+ EAPDME+L+PFV+SALPLAYGVLGVLLF
Sbjct: 424 LTIGSSVELGIASQINRLPPEVVKYFTDPDAVEAPDMELLFPFVDSALPLAYGVLGVLLF 603
Query: 181 HEVGHFLAAFPKQVKL 196
HEVGHFL+AFPKQ K+
Sbjct: 604 HEVGHFLSAFPKQGKI 651
>TC203794 weakly similar to UP|Q8YV67 (Q8YV67) Alr2114 protein, partial (15%)
Length = 1307
Score = 186 bits (473), Expect = 1e-47
Identities = 124/388 (31%), Positives = 192/388 (48%), Gaps = 1/388 (0%)
Frame = +2
Query: 23 GYSTFWVTKEEPFGELGEGILFIGNLRGKREDIFSILQNRLVEATGDKYNLFMVEEPDSD 82
G+ TF+VT ++P+ G+LF GNLRG+ + + RL + GD+Y LF++ P+ D
Sbjct: 2 GFDTFFVTSQDPYEG---GVLFKGNLRGQAAKSYDKISKRLKDKFGDEYKLFLLVNPEDD 172
Query: 83 SPDPRGGPRVSFGLLRKEVSEPEETTLWQYVVASLLFLLTIGTSVEVGIASQINRLPPEL 142
P PR + +PE T + ++ A L+T+ T + + L +L
Sbjct: 173 KPVAVVVPRTTL--------QPETTAVPEWFAAGSFGLITVFTL----LLRNVPALQSDL 316
Query: 143 VKFLTDPNYTEAPDMEMLYPFVESALPLAYGVLGVLLFHEVGHFLAAFPKQVKLSIPFFI 202
+ + N ++ LP A +L HE+GHFLAA VKL +P+F+
Sbjct: 317 LSTFDNLN------------LLKDGLPGALVTALILGVHELGHFLAAKDTGVKLGVPYFV 460
Query: 203 PHITLGSFGAITQFKSILPDRSTQVDISLAGPFAGAVLSFSMFAVGLLLSSNPDVAGDLV 262
P +GSFGAIT+ ++I+P+R + ++ AGP AG L + +G +L P G V
Sbjct: 461 PSWQIGSFGAITRIRNIVPNREDLLKVAAAGPIAGYALGLLLLLLGFVL---PPSDGIGV 631
Query: 263 QVPSMLFQGSLLLGLISRATLGYAAVHAATVPIHPLVIAGWCGLTIQAFNMLPVGCLDGG 322
V + +F S L G I++ LG + I+PLVI W GL I A N +P G LDGG
Sbjct: 632 VVDASVFHESFLAGGIAKLLLGNVLKEGTAISINPLVIWAWAGLLINAINSIPAGELDGG 811
Query: 323 RSVQGAFGKGATMVFGLTTYTLLGLGVLGGPLSLAWGFFVIFSQRSPEKPCLNDVTEVGT 382
R +G+ A++ F + LLG+ L ++ W + F QR P P ++T+ G
Sbjct: 812 RISFALWGRKASLRFTGVSIALLGVSSLLNDVAFYWVVLIFFLQRGPIAPLSEEITDPGE 991
Query: 383 WRQTFVGVAI-FLAVLTLLPVWDELAEE 409
+ +GV + L +L LP EE
Sbjct: 992 -KYVAIGVTVLLLGLLVCLPYPFPFTEE 1072
>CO983496
Length = 793
Score = 97.8 bits (242), Expect = 8e-21
Identities = 66/205 (32%), Positives = 95/205 (46%), Gaps = 11/205 (5%)
Frame = -2
Query: 208 GSFGAITQFKSILPDRSTQVDISLAGPFAGAVLSFSMFAVGLLLSSNPDVAGD-LVQVPS 266
G G + ++S+LP++ DI +A A A L+ + AV ++ GD + V
Sbjct: 780 GXXGVMNNYESLLPNKKALFDIPVART-ASAYLTSLLLAVAAFVADGSFNGGDNALYVRP 604
Query: 267 MLFQGSLLLGLISRATLGYA---------AVHAATVPIHPLVIAGWCGLTIQAFNMLPVG 317
F + LL I Y AV VP+ PL AG G+ + + NMLP G
Sbjct: 603 QFFYNNPLLSFIQYVIGPYTDDLGNVLPYAVEGVGVPVDPLAFAGLLGMVVTSLNMLPCG 424
Query: 318 CLDGGRSVQGAFGKGATMVFGLTTYTLLGLGVLGGP-LSLAWGFFVIFSQRSPEKPCLND 376
L+GGR Q FG+ M+ T LLG+G L G L LAWG F F + E P ++
Sbjct: 423 RLEGGRIAQAMFGRSTAMLLSFATSLLLGVGGLSGSVLCLAWGLFATFFRGGEEIPAKDE 244
Query: 377 VTEVGTWRQTFVGVAIFLAVLTLLP 401
++ +G R + V + LTL P
Sbjct: 243 ISPIGESRYAWGIVLGLICFLTLFP 169
>BE331202
Length = 490
Score = 84.7 bits (208), Expect = 7e-17
Identities = 38/52 (73%), Positives = 42/52 (80%)
Frame = +3
Query: 343 TLLGLGVLGGPLSLAWGFFVIFSQRSPEKPCLNDVTEVGTWRQTFVGVAIFL 394
TLLGLGVLGGPLSL WG +V+ QR+PEKPCLNDVTEVGTWR+ V FL
Sbjct: 330 TLLGLGVLGGPLSLPWGLYVLLCQRAPEKPCLNDVTEVGTWRKALVAWLFFL 485
>TC203872 weakly similar to UP|Q6NVM1 (Q6NVM1) MGC69430 protein, partial (4%)
Length = 550
Score = 42.7 bits (99), Expect = 3e-04
Identities = 28/92 (30%), Positives = 44/92 (47%), Gaps = 1/92 (1%)
Frame = +2
Query: 319 LDGGRSVQGAFGKGATMVFGLTTYTLLGLGVLGGPLSLAWGFFVIFSQRSPEKPCLNDVT 378
LDGGR +G+ A++ F + LLG+ L ++ W + F QR P P ++T
Sbjct: 95 LDGGRISFALWGRKASLRFTGVSIALLGVSSLLNDVAFYWVVLIFFLQRGPIAPLSEEIT 274
Query: 379 EVGTWRQTFVGVAI-FLAVLTLLPVWDELAEE 409
+ G + +GV + L +L LP EE
Sbjct: 275 DPGE-KYVAIGVTVLLLGLLVCLPYPFPFTEE 367
>BG839913 similar to GP|9955560|emb| protein kinase-like {Arabidopsis
thaliana}, partial (18%)
Length = 749
Score = 28.9 bits (63), Expect = 4.5
Identities = 25/81 (30%), Positives = 34/81 (41%), Gaps = 7/81 (8%)
Frame = -3
Query: 331 KGATMVFGLTTYTLLGLGV-------LGGPLSLAWGFFVIFSQRSPEKPCLNDVTEVGTW 383
+G T++ G T GV + L +A+ FF Q SP T
Sbjct: 690 RGRTVIIGFKRRTTSCFGVPESVQSFVASKLPMAFAFFFSL-QFSPHT--FASATARSLS 520
Query: 384 RQTFVGVAIFLAVLTLLPVWD 404
R F GV++ LAV T L +WD
Sbjct: 519 RSRFHGVSLVLAVFTGLSLWD 457
>TC216470 weakly similar to UP|Q9M6T9 (Q9M6T9) Lipid-transfer protein,
partial (61%)
Length = 648
Score = 28.1 bits (61), Expect = 7.7
Identities = 13/31 (41%), Positives = 15/31 (47%)
Frame = -2
Query: 290 AATVPIHPLVIAGWCGLTIQAFNMLPVGCLD 320
A P HPL I G GL + A N + C D
Sbjct: 332 AFEAPTHPLTIWGGFGLVVYALNPIAAWCRD 240
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.143 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,711,881
Number of Sequences: 63676
Number of extensions: 243007
Number of successful extensions: 1058
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 1053
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1056
length of query: 418
length of database: 12,639,632
effective HSP length: 100
effective length of query: 318
effective length of database: 6,272,032
effective search space: 1994506176
effective search space used: 1994506176
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137822.17


