

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137821.7 - phase: 0
(821 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
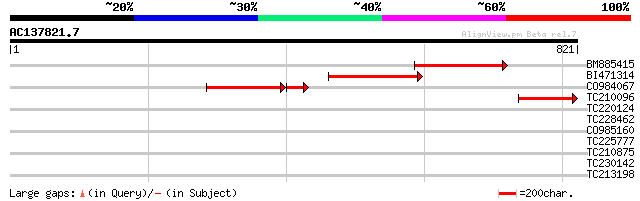
Score E
Sequences producing significant alignments: (bits) Value
BM885415 similar to SP|Q8RVQ5|SC10 Exocyst complex component Sec... 252 4e-67
BI471314 similar to SP|Q8RVQ5|SC10 Exocyst complex component Sec... 234 2e-61
CO984067 194 6e-60
TC210096 similar to UP|SC10_ARATH (Q8RVQ5) Exocyst complex compo... 160 2e-39
TC220124 weakly similar to UP|Q9LZU5 (Q9LZU5) Kinesin-related pr... 39 0.007
TC228462 similar to GB|AAQ56779.1|34098793|BT010336 At4g01400 {A... 31 1.9
CO985160 30 5.6
TC225777 weakly similar to UP|Q868B4 (Q868B4) C. elegans GRL-25 ... 29 7.4
TC210875 similar to UP|Q79FV6 (Q79FV6) PE-PGRS FAMILY PROTEIN, p... 29 9.6
TC230142 29 9.6
TC213198 similar to UP|Q9SY57 (Q9SY57) F14N23.3, partial (18%) 29 9.6
>BM885415 similar to SP|Q8RVQ5|SC10 Exocyst complex component Sec10.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (16%)
Length = 407
Score = 252 bits (644), Expect = 4e-67
Identities = 129/135 (95%), Positives = 134/135 (98%)
Frame = +1
Query: 586 ESSFRSFMVAVQRSGSSVAIIQQYFSNSISRLLLPVDGAHAAACEEMATAMSSAEAAAYK 645
ESSFRSFM+AVQRSGSSVAIIQQYF+NSISRLLLPVDGAHAAACEEMATAMSSAEAAAYK
Sbjct: 1 ESSFRSFMIAVQRSGSSVAIIQQYFANSISRLLLPVDGAHAAACEEMATAMSSAEAAAYK 180
Query: 646 GLQQCIETVMAEVERLLSAEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTA 705
GLQQCIETVMAEVERLLSAEQKATDY+SPDDGMAPDHR T+ACTRVVAYLSRVLESAFTA
Sbjct: 181 GLQQCIETVMAEVERLLSAEQKATDYRSPDDGMAPDHRATSACTRVVAYLSRVLESAFTA 360
Query: 706 LEGLNKQAFLSELGN 720
LEGLNKQAFL+ELGN
Sbjct: 361 LEGLNKQAFLTELGN 405
>BI471314 similar to SP|Q8RVQ5|SC10 Exocyst complex component Sec10.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (16%)
Length = 422
Score = 234 bits (596), Expect = 2e-61
Identities = 125/138 (90%), Positives = 134/138 (96%), Gaps = 1/138 (0%)
Frame = +2
Query: 462 ELRAESQ-ISDSSGTIGRSKGATVASSQQQISVTVVTEFVRWNEEAITRCNLFSSQPSTL 520
ELRAESQ ISD+SG+IGRSKGA+V SSQQQISVTVVTEFVRWNEEAI+RCNLF+SQP+TL
Sbjct: 8 ELRAESQQISDASGSIGRSKGASVVSSQQQISVTVVTEFVRWNEEAISRCNLFASQPATL 187
Query: 521 ATLVKAVFTCLLDQVSQYIAEGLERARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEA 580
AT VKAVFTCLLDQVSQYIA+GLERARD LTEAANLRERFVLGTSV+RRVAAAAASAAEA
Sbjct: 188 ATHVKAVFTCLLDQVSQYIADGLERARDSLTEAANLRERFVLGTSVTRRVAAAAASAAEA 367
Query: 581 AAAAGESSFRSFMVAVQR 598
AAAAGESSFRSFM+AVQR
Sbjct: 368 AAAAGESSFRSFMIAVQR 421
>CO984067
Length = 827
Score = 194 bits (494), Expect(2) = 6e-60
Identities = 101/115 (87%), Positives = 108/115 (93%)
Frame = -1
Query: 285 QFNRGTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQSSPNNVARGLSSLYKEITDTV 344
QFNRGTSAMQHYVATRPMFIDVE+MNADT+LVLGD A Q+SP+NVARGLSSLYKEITDTV
Sbjct: 749 QFNRGTSAMQHYVATRPMFIDVEIMNADTKLVLGDLAGQASPSNVARGLSSLYKEITDTV 570
Query: 345 RKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLVKPSLVNLPSMEEGGLL 399
KEAATITAVFPSP+EVMSILVQRVLEQ +TALLDKLL K SLVNLPS+EEG LL
Sbjct: 569 HKEAATITAVFPSPSEVMSILVQRVLEQ*ITALLDKLLEKSSLVNLPSVEEGELL 405
Score = 55.8 bits (133), Expect(2) = 6e-60
Identities = 25/32 (78%), Positives = 30/32 (93%)
Frame = -2
Query: 401 YLRMLAVSYEKTQEIARDLRTVGCGDLDVEGL 432
YLRMLAV+YEKT E+ARDL+ VGCGDLDV+G+
Sbjct: 301 YLRMLAVAYEKT*ELARDLQAVGCGDLDVDGV 206
>TC210096 similar to UP|SC10_ARATH (Q8RVQ5) Exocyst complex component Sec10,
partial (10%)
Length = 684
Score = 160 bits (405), Expect = 2e-39
Identities = 80/84 (95%), Positives = 84/84 (99%)
Frame = +3
Query: 738 SGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESLSTLFEGTPSIRKDA 797
SGGLRLKRDITEYGEF+RSFNAPSVDEKFELLGI+ANVFIVAPESLSTLFEGTPSIRKDA
Sbjct: 12 SGGLRLKRDITEYGEFLRSFNAPSVDEKFELLGIMANVFIVAPESLSTLFEGTPSIRKDA 191
Query: 798 QRFIQLREDYKSAKLASKLSSLWS 821
QRFIQLR+DYK+AKLASKLSSLWS
Sbjct: 192 QRFIQLRDDYKAAKLASKLSSLWS 263
>TC220124 weakly similar to UP|Q9LZU5 (Q9LZU5) Kinesin-related protein-like,
partial (26%)
Length = 864
Score = 39.3 bits (90), Expect = 0.007
Identities = 48/233 (20%), Positives = 103/233 (43%), Gaps = 13/233 (5%)
Frame = +3
Query: 75 ATSPLFPEVEKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGV- 133
AT L V KL +++ K L +L +++ D+ + +KH L +L KG+
Sbjct: 27 ATEDLRQRVGKLKNMYGSGIKALDDLAEELKVNNQLTYDDLKSEVAKHSSALEDLFKGIA 206
Query: 134 -------DGLFASFARLDSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNS 186
+ L +S + ++ +++ + H ++ + R + T+ + + S
Sbjct: 207 LEADSLLNDLQSSLHKQEANLTAYAHQQRE--SHARAVETTRAVSKITVNFFETIDRHAS 380
Query: 187 SPGDLMELSPLFSDDSRVAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAV 246
S +++E + L +D ++ E ++ ++ E+ + + A NA + +++AV
Sbjct: 381 SLTEIVEEAQLV-NDQKLCELEKKFEECTAYEEKQL-LEKVAEMLASSNARKKQLVQMAV 554
Query: 247 ANLQEYCN----ELENRLLSRFDAASQ-KRELTTMAECAKILSQFNRGTSAMQ 294
+L+E N +L L+ D+ S K E E K S ++ TSA++
Sbjct: 555 NDLRESANCRTSKLRQEALTMQDSTSSVKAEWRVHME--KTESNYHEDTSAVE 707
>TC228462 similar to GB|AAQ56779.1|34098793|BT010336 At4g01400 {Arabidopsis
thaliana;} , partial (45%)
Length = 1232
Score = 31.2 bits (69), Expect = 1.9
Identities = 46/209 (22%), Positives = 81/209 (38%), Gaps = 14/209 (6%)
Frame = +2
Query: 620 PVDGAHAAAC-EEMATAMSSAEAAAYKGLQQCIETVMAEVERLL------SAEQKATDYK 672
P D +C E+A + ++ + A G++Q + T+ + LL S E +Y
Sbjct: 392 PADREKVKSCLTELADSSNAFKQALNAGIEQLVATITPRIRPLLDSVGTISYELSEAEYA 571
Query: 673 SPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLHKVLLNHWQK 732
+ R A VA+L ++ + N F+ + + + K L +
Sbjct: 572 DNEVNDPWVQRLLYAVESNVAWLQPLMTAN-------NYDTFVHLIIDFIVKRLEVIMMQ 730
Query: 733 YTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESLSTLFEGTPS 792
F+ GGL+L RD +V +KF L +A + + S F G S
Sbjct: 731 KRFSQLGGLQLDRDARALVSHFSVMTQRTVRDKFARLTQMATILNLEKVSEILDFWGENS 910
Query: 793 -------IRKDAQRFIQLREDYKSAKLAS 814
+ +R + LR D+KS +A+
Sbjct: 911 GPMTWRLTPAEVRRVLGLRVDFKSEAIAA 997
>CO985160
Length = 777
Score = 29.6 bits (65), Expect = 5.6
Identities = 33/114 (28%), Positives = 48/114 (41%), Gaps = 3/114 (2%)
Frame = +2
Query: 566 VSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVAIIQQYFSNSISRLLLPVDGAH 625
V VAAA A AEAAA A + + VA + V + A
Sbjct: 392 VEVEVAAAVAGFAEAAAVAAAQAAAAGPVAAAVAAEEVVAAAAAAEEVV---------AA 544
Query: 626 AAACEEMATAMSSAE--AAAYKGLQQCIETVM-AEVERLLSAEQKATDYKSPDD 676
AAA EE+ A+ AE AAA ++ + V+ AE +++ E A + +D
Sbjct: 545 AAAAEEVVAAVVGAEEVAAAAAAAEEVVAAVVGAEEVAVVACEVAAAVVATAED 706
>TC225777 weakly similar to UP|Q868B4 (Q868B4) C. elegans GRL-25 protein
(Corresponding sequence ZK643.8), partial (7%)
Length = 762
Score = 29.3 bits (64), Expect = 7.4
Identities = 26/81 (32%), Positives = 39/81 (48%)
Frame = +2
Query: 563 GTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVAIIQQYFSNSISRLLLPVD 622
G + + +AAAA+A AAAAA + + + + ++ I S SIS +P
Sbjct: 140 GAATTTATSAAAAAATTAAAAAATPAATTATPSTTTTAATATDISANIS-SISSAAVPPS 316
Query: 623 GAHAAACEEMATAMSSAEAAA 643
A AA ATA ++A A A
Sbjct: 317 PAFAAV---PATAAAAATATA 370
>TC210875 similar to UP|Q79FV6 (Q79FV6) PE-PGRS FAMILY PROTEIN, partial (3%)
Length = 649
Score = 28.9 bits (63), Expect = 9.6
Identities = 16/52 (30%), Positives = 28/52 (53%)
Frame = +1
Query: 563 GTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVAIIQQYFSNSI 614
G+ R+A + A AAAAA +S R + +RS S++ +++FS +
Sbjct: 58 GSETRTRIAGSQARGPPAAAAAANASGRRSWQSFRRSRISISG*RRFFSQKV 213
>TC230142
Length = 901
Score = 28.9 bits (63), Expect = 9.6
Identities = 16/48 (33%), Positives = 26/48 (53%)
Frame = +1
Query: 360 EVMSILVQRVLEQRVTALLDKLLVKPSLVNLPSMEEGGLLFYLRMLAV 407
E +L Q +L ++VT + L VKP + +PS +GG+ L + V
Sbjct: 271 ECHKVLQQNMLRKKVTRQQNWLPVKPGAL*VPSSLQGGIFLKLFTMVV 414
>TC213198 similar to UP|Q9SY57 (Q9SY57) F14N23.3, partial (18%)
Length = 596
Score = 28.9 bits (63), Expect = 9.6
Identities = 19/63 (30%), Positives = 32/63 (50%), Gaps = 2/63 (3%)
Frame = +1
Query: 757 FNAPSVDEKFELLGILANVFIVAPESLSTLFEGTPSIRKDAQRFIQLREDY--KSAKLAS 814
++APSV K + L L + F+ ESLS + + +D + F++ D S K S
Sbjct: 241 YHAPSVARKRKRLSKLLSAFVSVAESLSESADTIGIVSRDVKVFLESDSDQIPNSLKQIS 420
Query: 815 KLS 817
K++
Sbjct: 421 KIA 429
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.129 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,313,766
Number of Sequences: 63676
Number of extensions: 285624
Number of successful extensions: 1610
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1511
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1568
length of query: 821
length of database: 12,639,632
effective HSP length: 105
effective length of query: 716
effective length of database: 5,953,652
effective search space: 4262814832
effective search space used: 4262814832
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC137821.7


