

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137821.5 - phase: 0
(589 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
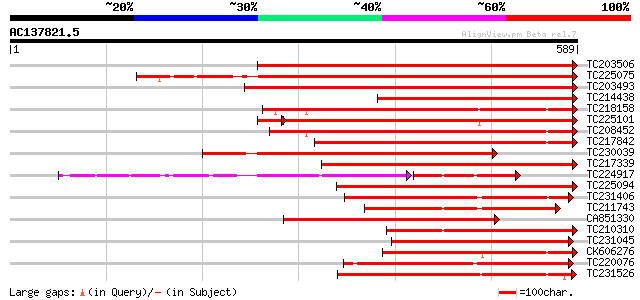
Score E
Sequences producing significant alignments: (bits) Value
TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (... 410 e-115
TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 399 e-111
TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like prote... 396 e-110
TC214438 similar to UP|Q708X4 (Q708X4) Pectin methylesterase (F... 385 e-107
TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pe... 364 e-101
TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 346 e-100
TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (... 343 9e-95
TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 343 2e-94
TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase i... 334 6e-92
TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase p... 329 2e-90
TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4... 219 2e-88
TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 288 6e-78
TC231406 similar to UP|Q9FF78 (Q9FF78) Pectinesterase, partial (... 265 6e-71
TC211743 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 264 9e-71
CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.... 264 9e-71
TC210310 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 251 6e-67
TC231045 similar to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase iso... 243 2e-64
CK606276 241 9e-64
TC220076 homologue to UP|PME3_PHAVU (Q43111) Pectinesterase 3 pr... 236 2e-62
TC231526 similar to UP|PME_MEDSA (Q42920) Pectinesterase precurs... 220 2e-57
>TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (57%)
Length = 1320
Score = 410 bits (1055), Expect = e-115
Identities = 197/335 (58%), Positives = 250/335 (73%), Gaps = 3/335 (0%)
Frame = +2
Query: 258 WPEWISAGDRRLLQG--STVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIKIKAGV 315
+P W++ DRRLL S ++AD+ V+ G+G KT++EA+ AP S++R++I ++AG
Sbjct: 11 FPGWLNKRDRRLLSLPLSAIQADITVSKTGNGTVKTIAEAIKKAPEHSTRRFIIYVRAGR 190
Query: 316 YKEN-VEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQ 374
Y+EN ++V +KKTN+MF+GDG+ T+ITG +NV+DG TTFHSA+ A G F+ARDITF+
Sbjct: 191 YEENNLKVGRKKTNVMFIGDGKGKTVITGKKNVIDGMTTFHSASFAASGAGFIARDITFE 370
Query: 375 NTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSA 434
N AGPAKHQAVALRVGAD + Y C+I+ YQD+ YVH+NRQFF C I GTVDFIFGN+A
Sbjct: 371 NYAGPAKHQAVALRVGADHAVVYRCNIVGYQDSCYVHSNRQFFRECNIYGTVDFIFGNAA 550
Query: 435 VVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLG 494
VVFQ C+I+AR+P + QKN +TAQ R DPNQNTGI + CRI DL VKG+FPTYLG
Sbjct: 551 VVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISLHNCRILPAPDLAPVKGSFPTYLG 730
Query: 495 RPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGF 554
RPWK+YSRTV++ S + D I P GW EWNG+FALNTL Y EY N GPGA +RV W G+
Sbjct: 731 RPWKQYSRTVYLVSYMGDHIHPRGWLEWNGDFALNTLYYGEYMNYGPGAAVGQRVQWPGY 910
Query: 555 KVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
+VI S EA FT FI GS+WL STG F+ GL
Sbjct: 911 RVIKSTMEANRFTVAQFISGSAWLPSTGVAFAAGL 1015
>TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (87%)
Length = 1744
Score = 399 bits (1025), Expect = e-111
Identities = 220/462 (47%), Positives = 282/462 (60%), Gaps = 4/462 (0%)
Frame = +1
Query: 132 REKIALHDCLETIDETLDELKEA----QNDLVLYPSKKTLYQHADDLKTLISSAITNQVT 187
R A+ DCL+ +D + D L A QN + S L + DL+T +S+A+ + T
Sbjct: 238 RLSTAIADCLDLLDLSSDVLSWALSASQNPKGKHNSTGNL---SSDLRTWLSAALAHPET 408
Query: 188 CLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNR 247
C++GF + + V+ ++ G V + LA D+ A S+K +
Sbjct: 409 CMEGF--EGTNSIVKGLVSAGIGQVVSLVEQLLAQVLPAQDQFDA---------ASSKGQ 555
Query: 248 KLLEEENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRY 307
+P WI +R+LLQ V DV VA DGSGN+ + +AV AAP S KR+
Sbjct: 556 ----------FPSWIKPKERKLLQAIAVTPDVTVALDGSGNYAKIMDAVLAAPDYSMKRF 705
Query: 308 VIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFL 367
VI +K GVY ENVE+ KKK NIM LG G T+I+G+R+VVDG TTF SAT A+ G F+
Sbjct: 706 VILVKKGVYVENVEIKKKKWNIMILGQGMDATVISGNRSVVDGWTTFRSATFAVSGRGFI 885
Query: 368 ARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVD 427
ARDI+FQNTAGP KHQAVALR +DLS F+ C I YQD+LY H RQFF +C ISGTVD
Sbjct: 886 ARDISFQNTAGPEKHQAVALRSDSDLSVFFRCGIFGYQDSLYTHTMRQFFRDCTISGTVD 1065
Query: 428 FIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKG 487
+IFG++ VFQNC + ++ QKN +TA GR DPN+ TG Q C I A DL G
Sbjct: 1066YIFGDATAVFQNCFLRVKKGLPNQKNTITAHGRKDPNEPTGFSFQFCNITADSDLIPSVG 1245
Query: 488 NFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSK 547
TYLGRPWK YSRTVFMQS +S+VI GW EWNGNFAL+TL Y EY NTG GAG +
Sbjct: 1246TAQTYLGRPWKSYSRTVFMQSYMSEVIGAEGWLEWNGNFALDTLYYAEYMNTGAGAGVAN 1425
Query: 548 RVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
RV W G+ + +++A +FT FI G+ WL STG F+ GL
Sbjct: 1426RVKWPGYHALNDSSQASNFTVSQFIEGNLWLPSTGVTFTAGL 1551
>TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like protein
(At3g49220), partial (59%)
Length = 1298
Score = 396 bits (1018), Expect = e-110
Identities = 196/349 (56%), Positives = 248/349 (70%), Gaps = 4/349 (1%)
Frame = +1
Query: 245 KNRKLLEEENGVGWPEWISAGDRRLLQG--STVKADVVVAADGSGNFKTVSEAVAAAPLK 302
+NR+ L +P W++ DRRLL S ++AD+VV+ DG+G KT++EA+ P
Sbjct: 100 QNRRRLMAMREDNFPTWLNGRDRRLLSLPLSQIQADIVVSKDGNGTVKTIAEAIKKVPEY 279
Query: 303 SSKRYVIKIKAGVYKE-NVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAI 361
SS+R +I I+AG Y+E N+++ +KKTN+MF+GDG+ T+ITG RN TTFH+A+ A
Sbjct: 280 SSRRIIIYIRAGRYEEDNLKLGRKKTNVMFIGDGKGKTVITGGRNYYQNLTTFHTASFAA 459
Query: 362 VGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCF 421
G F+A+D+TF+N AGP +HQAVALRVGAD + Y C+II YQDT+YVH+NRQF+ C
Sbjct: 460 SGSGFIAKDMTFENYAGPGRHQAVALRVGADHAVVYRCNIIGYQDTMYVHSNRQFYRECD 639
Query: 422 ISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKD 481
I GTVDFIFGN+AVVFQNC + AR+P + QKN +TAQ R DPNQNTGI I CRI AT D
Sbjct: 640 IYGTVDFIFGNAAVVFQNCTLWARKPMAQQKNTITAQNRKDPNQNTGISIHNCRIMATPD 819
Query: 482 LEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWN-GNFALNTLVYREYQNTG 540
LE KG++PTYLGRPWK Y+RTVFM S I D + P GW EWN +FAL+T Y EY N G
Sbjct: 820 LEASKGSYPTYLGRPWKLYARTVFMLSYIGDHVHPRGWLEWNTSSFALDTCYYGEYMNYG 999
Query: 541 PGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
PG+ +RV W G++ I S EA FT G FI GSSWL STG F GL
Sbjct: 1000PGSALGQRVNWAGYRAINSTVEASRFTVGQFISGSSWLPSTGVAFIAGL 1146
>TC214438 similar to UP|Q708X4 (Q708X4) Pectin methylesterase (Fragment) ,
partial (49%)
Length = 1026
Score = 385 bits (990), Expect = e-107
Identities = 181/207 (87%), Positives = 194/207 (93%)
Frame = +3
Query: 383 QAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDI 442
QAVALRVG DLSAF+NCDI+A+QDTLYVHNNRQFF C I+GTVDFIFGNSAVVFQ+CDI
Sbjct: 6 QAVALRVGGDLSAFFNCDILAFQDTLYVHNNRQFFEKCLIAGTVDFIFGNSAVVFQDCDI 185
Query: 443 HARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSR 502
HAR P+SGQKNMVTAQGRVDPNQNTGIVIQKCRIGAT DLE VK NF TYLGRPWKEYSR
Sbjct: 186 HARLPSSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATNDLESVKKNFKTYLGRPWKEYSR 365
Query: 503 TVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAE 562
TV MQSSISDVIDP+GWHEW+GNFAL+TLVYREYQNTGPGAGTS RVTWKG+KVIT AAE
Sbjct: 366 TVIMQSSISDVIDPIGWHEWSGNFALSTLVYREYQNTGPGAGTSNRVTWKGYKVITDAAE 545
Query: 563 AQSFTPGNFIGGSSWLGSTGFPFSLGL 589
A+ +TPG+FIGGSSWLGSTGFPFSLGL
Sbjct: 546 ARDYTPGSFIGGSSWLGSTGFPFSLGL 626
>TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pectin
methylesterase-like protein, partial (59%)
Length = 1291
Score = 364 bits (935), Expect = e-101
Identities = 188/335 (56%), Positives = 233/335 (69%), Gaps = 8/335 (2%)
Frame = +3
Query: 263 SAGDRRLLQGST-----VKADVVVAADGSGNFKTVSEAVAAAPLKSSKR---YVIKIKAG 314
S R+LLQ V+ V V+ DGSGNF T+++A+AAAP KS ++I + AG
Sbjct: 90 SVSRRKLLQAKVGDEVVVRDIVTVSQDGSGNFTTINDAIAAAPNKSVSTDGYFLIYVTAG 269
Query: 315 VYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQ 374
VY+ENV + KKKT +M +GDG TIITG+R+VVDG TTF SAT+A+VG F+ ++T +
Sbjct: 270 VYEENVSIDKKKTYLMMVGDGINKTIITGNRSVVDGWTTFSSATLAVVGQGFVGVNMTIR 449
Query: 375 NTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSA 434
NTAG KHQAVALR GADLS FY+C YQDTLYVH+ RQF+ C I GTVDFIFGN+
Sbjct: 450 NTAGAVKHQAVALRSGADLSTFYSCSFEGYQDTLYVHSLRQFYSECDIYGTVDFIFGNAK 629
Query: 435 VVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLG 494
VVFQNC ++ R P SGQ N +TAQGR DPNQ+TGI I C I A DL G TYLG
Sbjct: 630 VVFQNCKMYPRLPMSGQFNAITAQGRTDPNQDTGISIHNCTIRAADDLAASNG-VATYLG 806
Query: 495 RPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGF 554
RPWKEYSRTV+MQ+ + VI GW EW+G+FAL+TL Y EY N+GPG+GT RVTW G+
Sbjct: 807 RPWKEYSRTVYMQTVMDSVIHAKGWREWDGDFALSTLYYAEYSNSGPGSGTDNRVTWPGY 986
Query: 555 KVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
VI +A +A +FT NF+ G WL TG ++ L
Sbjct: 987 HVI-NATDAANFTVSNFLLGDDWLPQTGVSYTNNL 1088
>TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (64%)
Length = 1177
Score = 346 bits (888), Expect(2) = e-100
Identities = 174/308 (56%), Positives = 214/308 (68%), Gaps = 3/308 (0%)
Frame = +3
Query: 285 GSGNFKTVSEAVAAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGS 344
G N+ V +AV AAP S +RYVI IK GVY ENVE+ KKK N+M +GDG TII+G+
Sbjct: 207 GRENYTKVMDAVLAAPNYSMQRYVIHIKRGVYYENVEIKKKKWNLMMVGDGMDATIISGN 386
Query: 345 RNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAY 404
R+ +DG TTF SAT A+ G F+ARDITFQNTAGP KHQAVALR +DLS F+ C I Y
Sbjct: 387 RSFIDGWTTFRSATFAVSGRGFIARDITFQNTAGPEKHQAVALRSDSDLSVFFRCGIFGY 566
Query: 405 QDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPN 464
QD+LY H RQF+ C ISGTVDFIFG++ +FQNC I A++ QKN +TA GR +P+
Sbjct: 567 QDSLYTHTMRQFYRECKISGTVDFIFGDATAIFQNCHISAKKGLPNQKNTITAHGRKNPD 746
Query: 465 QNTGIVIQKCRIGATKDLEGVK---GNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHE 521
+ TG IQ C I A DL + TYLGRPWK YSRT+FMQS ISDV+ P GW E
Sbjct: 747 EPTGFSIQFCNISADYDLVNSVNSCNSTHTYLGRPWKPYSRTIFMQSYISDVLRPEGWLE 926
Query: 522 WNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGST 581
WNG+FAL+TL Y EY N GPGAG + RV W+G+ V+ +++A +FT FI G+ L ST
Sbjct: 927 WNGDFALDTLYYAEYMNYGPGAGVANRVKWQGYHVMNDSSQASNFTVSQFIEGNLCLPST 1106
Query: 582 GFPFSLGL 589
G F+ GL
Sbjct: 1107GVTFTAGL 1130
Score = 39.7 bits (91), Expect(2) = e-100
Identities = 15/30 (50%), Positives = 21/30 (70%)
Frame = +2
Query: 258 WPEWISAGDRRLLQGSTVKADVVVAADGSG 287
+P W+ G+R+LLQ + V D VV ADG+G
Sbjct: 125 YPSWVKTGERKLLQANVVSFDAVVTADGTG 214
>TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (56%)
Length = 1273
Score = 343 bits (881), Expect = 9e-95
Identities = 170/322 (52%), Positives = 224/322 (68%), Gaps = 3/322 (0%)
Frame = +1
Query: 271 QGSTVKADVVVAADGSGNFKTVSEAVAAAPLKSSKR---YVIKIKAGVYKENVEVPKKKT 327
QG + V+V+ G N+ ++ +A+AAAP + +++ ++ G+Y+E V +PK+K
Sbjct: 268 QGILLYDFVIVSHYGIDNYTSIGDAIAAAPNNTKPEDGYFLVYVREGLYEEYVVIPKEKK 447
Query: 328 NIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVAL 387
NI+ +GDG TIITG+ +V+DG TTF+S+T A+ G F+A D+TF+NTAGP KHQAVA+
Sbjct: 448 NILLVGDGINKTIITGNHSVIDGWTTFNSSTFAVSGERFIAVDVTFRNTAGPEKHQAVAV 627
Query: 388 RVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRP 447
R ADLS FY C YQDTLYVH+ RQF+ C I GTVDFIFGN+AVVFQ C I+AR+P
Sbjct: 628 RNNADLSTFYRCSFEGYQDTLYVHSLRQFYRECEIYGTVDFIFGNAAVVFQGCKIYARKP 807
Query: 448 NSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQ 507
QKN VTAQGR DPNQNTGI IQ C I A DL + ++LGRPWK YSRTV++Q
Sbjct: 808 LPNQKNAVTAQGRTDPNQNTGISIQNCSIDAAPDLVADLNSTMSFLGRPWKVYSRTVYLQ 987
Query: 508 SSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFT 567
S I +VI P GW EWNG L+TL Y E+ N GPG+ TS RVTW G+ ++ +A +A +FT
Sbjct: 988 SYIGNVIQPAGWLEWNGTVGLDTLFYGEFNNYGPGSNTSNRVTWPGYSLL-NATQAWNFT 1164
Query: 568 PGNFIGGSSWLGSTGFPFSLGL 589
NF G++WL T P++ GL
Sbjct: 1165VLNFTLGNTWLPDTDIPYTEGL 1230
>TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (48%)
Length = 1225
Score = 343 bits (879), Expect = 2e-94
Identities = 161/273 (58%), Positives = 202/273 (73%)
Frame = +3
Query: 317 KENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNT 376
KEN+++ + N+M +GDG TI+TG+ N +DGSTTF SAT A+ G F+ARDITF+NT
Sbjct: 6 KENIDIKRTVKNLMIVGDGMGATIVTGNHNAIDGSTTFRSATFAVDGDGFIARDITFENT 185
Query: 377 AGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVV 436
AGP KHQAVALR GAD S FY C YQDTLYV+ NRQF+ +C I GTVDFIFG++ V
Sbjct: 186 AGPQKHQAVALRSGADHSVFYRCSFRGYQDTLYVYANRQFYRDCDIYGTVDFIFGDAVAV 365
Query: 437 FQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRP 496
QNC+I+ R+P S Q+N VTAQGR DPN+NTGI+I CRI A DL+ V+G+F T+LGRP
Sbjct: 366 LQNCNIYVRKPMSNQQNTVTAQGRTDPNENTGIIIHNCRITAAGDLKAVQGSFRTFLGRP 545
Query: 497 WKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKV 556
W++YSRTV M+S++ +I P GW W+GNFAL+TL Y E+ NTG GA T RV W GF+V
Sbjct: 546 WQKYSRTVVMKSALDGLISPAGWFPWSGNFALSTLYYAEHANTGAGASTGGRVDWAGFRV 725
Query: 557 ITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
I S+ EA FT GNF+ G SW+ +G PF GL
Sbjct: 726 I-SSTEAVKFTVGNFLAGGSWIPGSGVPFDEGL 821
>TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (70%)
Length = 913
Score = 334 bits (857), Expect = 6e-92
Identities = 169/310 (54%), Positives = 219/310 (70%), Gaps = 4/310 (1%)
Frame = +1
Query: 201 VRKVLQEGQIHVEHMCSNALAMTKNMTD--KDIAEFEQTNMVLGSNKNRKLLEEENGVGW 258
VR +EG + + H SN+LAM K + K +A + G K+ G+
Sbjct: 13 VRDRFEEGLLEISHHVSNSLAMLKKLPAGVKKLASKNEVFPGYGKIKD----------GF 162
Query: 259 PEWISAGDRRLLQGSTVKAD--VVVAADGSGNFKTVSEAVAAAPLKSSKRYVIKIKAGVY 316
P W+S DR+LLQ + + + ++VA DG+GNF T++EAVA AP S+ R+VI IKAG Y
Sbjct: 163 PTWLSTKDRKLLQAAVNETNFNLLVAKDGTGNFTTIAEAVAVAPNSSATRFVIHIKAGAY 342
Query: 317 KENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNT 376
ENVEV +KKTN+MF+GDG T++ SRNVVDG TTF SATVA+VG F+A+ ITF+N+
Sbjct: 343 FENVEVIRKKTNLMFVGDGIGKTVVKASRNVVDGWTTFQSATVAVVGDGFIAKGITFENS 522
Query: 377 AGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVV 436
AGP+KHQAVALR G+D SAFY C +AYQDTLYVH+ RQF+ +C + GTVDFIFGN+A V
Sbjct: 523 AGPSKHQAVALRSGSDFSAFYKCSFVAYQDTLYVHSLRQFYRDCDVYGTVDFIFGNAATV 702
Query: 437 FQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRP 496
QNC+++AR+PN Q+N+ TAQGR DPNQNTGI I C++ A DL VK F YLGRP
Sbjct: 703 LQNCNLYARKPNENQRNLFTAQGREDPNQNTGISILNCKVAAAADLIPVKSQFKNYLGRP 882
Query: 497 WKEYSRTVFM 506
WK+YSRTV++
Sbjct: 883 WKKYSRTVYL 912
>TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precursor,
partial (48%)
Length = 923
Score = 329 bits (843), Expect = 2e-90
Identities = 161/267 (60%), Positives = 195/267 (72%), Gaps = 2/267 (0%)
Frame = +1
Query: 325 KKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQA 384
+KTN+M +GDG+ T+ITG+ N +DG+TTF +ATVA VG F+A+DI FQNTAGP KHQA
Sbjct: 1 EKTNVMLVGDGKDATVITGNLNFIDGTTTFKTATVAAVGDGFIAQDIWFQNTAGPQKHQA 180
Query: 385 VALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHA 444
VALRVGAD S C I A+QDTLY H+NRQF+ + FI+GTVDFIFGN+AVVFQ CD+ A
Sbjct: 181 VALRVGADQSVINRCRIDAFQDTLYAHSNRQFYRDSFITGTVDFIFGNAAVVFQKCDLVA 360
Query: 445 RRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTV 504
R+P Q NMVTAQGR DPNQNTG IQ+C + + DL+ V G+ T+LGRPWK+YSRTV
Sbjct: 361 RKPMDKQNNMVTAQGREDPNQNTGTSIQQCNLTPSSDLKPVVGSIKTFLGRPWKKYSRTV 540
Query: 505 FMQSSISDVIDPVGWHEWNGNFA--LNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAE 562
MQS++ IDP GW EW+ L TL Y EY N GPGAGTSKRV W G+ +I +AAE
Sbjct: 541 VMQSTLDSHIDPTGWAEWDAQSKDFLQTLYYGEYMNNGPGAGTSKRVNWPGYHIIKTAAE 720
Query: 563 AQSFTPGNFIGGSSWLGSTGFPFSLGL 589
A FT I G+ WL +TG F GL
Sbjct: 721 ASKFTVAQLIQGNVWLKNTGVNFIEGL 801
>TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4L23.27
{Arabidopsis thaliana;} , partial (69%)
Length = 1483
Score = 219 bits (558), Expect(2) = 2e-88
Identities = 145/369 (39%), Positives = 198/369 (53%), Gaps = 2/369 (0%)
Frame = +2
Query: 51 NSIASSSLSLSHHSHAILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITT 110
+SIASS +S ++S C T YP+ C +++ I + D + +SL +
Sbjct: 167 SSIASS------YSFKDIQSWCNQTPYPQPCEYYLTNHA-FNKPIKSKSDFLKVSLQLAL 325
Query: 111 RAVEHNYFTVEKLLLRKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQH 170
+ + L K EK A DCL+ + T+ L + N P+ K +
Sbjct: 326 ERAQRSELNTHALG-PKCRNVHEKAAWADCLQLYEYTIQRLNKTIN-----PNTKC---N 478
Query: 171 ADDLKTLISSAITNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKD 230
D +T +S+A+TN TC +GF V ++ +V + SN L++ K
Sbjct: 479 QTDTQTWLSTALTNLETCKNGFYELGVPDYVLPLMSN---NVTKLLSNTLSLNKGPYQYK 649
Query: 231 IAEFEQTNMVLGSNKNRKLLEEENGVGWPEWISAGDRRLLQGSTV--KADVVVAADGSGN 288
+++ G+P W+ GDR+LLQ S+V A+VVVA DGSG
Sbjct: 650 PPSYKE--------------------GFPTWVKPGDRKLLQSSSVASNANVVVAKDGSGK 769
Query: 289 FKTVSEAVAAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVV 348
+ TV AV AAP SS RYVI +K+GVY E VEV K NIM +GDG TIITGS++V
Sbjct: 770 YTTVKAAVDAAPKSSSGRYVIYVKSGVYNEQVEV--KGNNIMLVGDGIGKTIITGSKSVG 943
Query: 349 DGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTL 408
G+TTF SATVA VG F+A+DITF+NTAG A HQAVA R G+DLS FY C +QDTL
Sbjct: 944 GGTTTFRSATVAAVGDGFIAQDITFRNTAGAANHQAVAFRSGSDLSVFYRCSFEGFQDTL 1123
Query: 409 YVHNNRQFF 417
YVH+ RQF+
Sbjct: 1124YVHSERQFY 1150
Score = 125 bits (315), Expect(2) = 2e-88
Identities = 60/111 (54%), Positives = 78/111 (70%)
Frame = +3
Query: 420 CFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGAT 479
C I GTVDFIFGN+A V QNC+I+AR P + VTAQGR DPNQNTGI+I ++
Sbjct: 1158 CDIYGTVDFIFGNAAAVLQNCNIYARTPPQ-RTITVTAQGRTDPNQNTGIIIHNSKVTGA 1334
Query: 480 KDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNT 530
+ +YLGRPW++YSRTVFM++ + +I+P GW EW+GNFAL+T
Sbjct: 1335 SGFN--PSSVKSYLGRPWQKYSRTVFMKTYLDSLINPAGWMEWDGNFALDT 1481
>TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (47%)
Length = 919
Score = 288 bits (736), Expect = 6e-78
Identities = 141/250 (56%), Positives = 170/250 (67%)
Frame = +3
Query: 340 IITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNC 399
II+G+R+VVDG TTF SAT A+ G F+ARDI+FQNTAGP KHQAVALR DLS F+ C
Sbjct: 3 IISGNRSVVDGWTTFRSATFAVSGRGFIARDISFQNTAGPEKHQAVALRSDTDLSVFFRC 182
Query: 400 DIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQG 459
I YQD+LY H RQFF C I+GTVD+IFG++ VFQNC + ++ QKN +TA G
Sbjct: 183 GIFGYQDSLYTHTMRQFFRECTITGTVDYIFGDATAVFQNCFLRVKKGLPNQKNTITAHG 362
Query: 460 RVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGW 519
R DPN+ TG Q C I A DL + +YLGRPWK YSRTVFMQS +S+VI GW
Sbjct: 363 RKDPNEPTGFSFQFCNITADSDLVPWVSSTQSYLGRPWKSYSRTVFMQSYMSEVIRGEGW 542
Query: 520 HEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLG 579
EWNGNFAL TL Y EY NTG GAG + RV W G+ + +A +FT FI G+ WL
Sbjct: 543 LEWNGNFALETLYYGEYMNTGAGAGLANRVKWPGYHPFNDSNQASNFTVAQFIEGNLWLP 722
Query: 580 STGFPFSLGL 589
STG ++ GL
Sbjct: 723 STGVTYTAGL 752
>TC231406 similar to UP|Q9FF78 (Q9FF78) Pectinesterase, partial (35%)
Length = 851
Score = 265 bits (676), Expect = 6e-71
Identities = 133/238 (55%), Positives = 159/238 (65%)
Frame = +2
Query: 348 VDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDT 407
VDG+ TF +AT A+ G NF+ARD+ F+NTAGP K QAVAL AD + +Y C I A+QD+
Sbjct: 5 VDGTPTFSTATFAVFGRNFIARDMGFRNTAGPQKQQAVALMTSADQAVYYRCQIDAFQDS 184
Query: 408 LYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNT 467
LY H+NRQF+ C I GTVDFIFGNSAVV QNC+I R P GQ+N +TAQG+ DPN NT
Sbjct: 185 LYAHSNRQFYRECNIYGTVDFIFGNSAVVLQNCNIMPRVPMQGQQNPITAQGKTDPNMNT 364
Query: 468 GIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFA 527
GI IQ C I DL VK TYLGRPWK YS VFM S + I P GW W GN A
Sbjct: 365 GISIQNCNITPFGDLSSVK----TYLGRPWKNYSTPVFM*SPMGIFIHPNGWLPWVGNSA 532
Query: 528 LNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPF 585
+T+ Y E+QN GPGA T RV WKG +VIT +A FT F+ G W+ ++G PF
Sbjct: 533 PDTIFYAEFQNVGPGASTKNRVNWKGLRVIT-RKQASMFTVKAFLSGERWITASGAPF 703
>TC211743 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (39%)
Length = 601
Score = 264 bits (674), Expect = 9e-71
Identities = 127/204 (62%), Positives = 156/204 (76%)
Frame = +2
Query: 369 RDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDF 428
+DITF+NTAG HQAVALR G+DLS FY C YQDTLYV+++RQF+ C I GTVDF
Sbjct: 2 QDITFRNTAGATNHQAVALRSGSDLSVFYRCSFEGYQDTLYVYSDRQFYRECDIYGTVDF 181
Query: 429 IFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGN 488
IFGN+AVVFQNC+I+AR P + + N +TAQGR DPNQNTGI I ++ A DL GV+
Sbjct: 182 IFGNAAVVFQNCNIYARNPPN-KVNTITAQGRTDPNQNTGISIHNSKVTAASDLMGVR-- 352
Query: 489 FPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKR 548
TYLGRPW++YSRTVFM++ + +I+P GW EW+GNFAL+TL Y EY NTGPG+ T+ R
Sbjct: 353 --TYLGRPWQQYSRTVFMKTYLDSLINPEGWLEWSGNFALSTLYYGEYMNTGPGSSTANR 526
Query: 549 VTWKGFKVITSAAEAQSFTPGNFI 572
V W G+ VITSA+EA FT GNFI
Sbjct: 527 VNWLGYHVITSASEASKFTVGNFI 598
>CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.150 -
Arabidopsis thaliana, partial (37%)
Length = 700
Score = 264 bits (674), Expect = 9e-71
Identities = 136/225 (60%), Positives = 163/225 (72%), Gaps = 1/225 (0%)
Frame = +2
Query: 285 GSGNFKTVSEAV-AAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITG 343
GSGNFKTV +A+ AAA K R+VI +K GVY+EN+EV NIM +GDG NTIIT
Sbjct: 14 GSGNFKTVQDALNAAAKRKEKTRFVIHVKKGVYRENIEVALHNDNIMLVGDGLRNTIITS 193
Query: 344 SRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIA 403
+R+V DG TT+ SAT I G +F+ARDITFQN+AG K QAVALR +DLS FY C I+
Sbjct: 194 ARSVQDGYTTYSSATAGIDGLHFIARDITFQNSAGVHKGQAVALRSASDLSVFYRCGIMG 373
Query: 404 YQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDP 463
YQDTL H RQF+ C+I GTVDFIFGN+AVVFQNC I ARRP GQ NM+TAQGR DP
Sbjct: 374 YQDTLMAHAQRQFYRQCYIYGTVDFIFGNAAVVFQNCYIFARRPLEGQANMITAQGRGDP 553
Query: 464 NQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQS 508
QNTGI I +I A DL+ V + T+LGRPW++YSR V M++
Sbjct: 554 FQNTGISIHNSQIRAAPDLKPVVDKYNTFLGRPWQQYSRVVVMKT 688
>TC210310 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (38%)
Length = 678
Score = 251 bits (641), Expect = 6e-67
Identities = 118/198 (59%), Positives = 146/198 (73%)
Frame = +1
Query: 392 DLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQ 451
DLS FY C YQDTLYVH+ RQF+ C I GTVDFIFGN+AVV QNC+I AR P + +
Sbjct: 1 DLSVFYKCSFEGYQDTLYVHSERQFYRECNIYGTVDFIFGNAAVVLQNCNIFARNPPN-K 177
Query: 452 KNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSIS 511
N +TAQGR DPNQNTGI I R+ A DL V+ + TYLGRPWK+YSRTVFM++ +
Sbjct: 178 VNTITAQGRTDPNQNTGISIHNSRVTAASDLRPVQNSVRTYLGRPWKQYSRTVFMKTYLD 357
Query: 512 DVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNF 571
+I+P GW EW+GNFAL+TL Y EY NTGPG+ T++RV W G++VITSA+EA F+ NF
Sbjct: 358 GLINPAGWMEWSGNFALDTLYYGEYMNTGPGSSTARRVKWSGYRVITSASEASKFSVANF 537
Query: 572 IGGSSWLGSTGFPFSLGL 589
I G++WL ST PF+ L
Sbjct: 538 IAGNAWLPSTKVPFTPSL 591
>TC231045 similar to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (69%)
Length = 747
Score = 243 bits (619), Expect = 2e-64
Identities = 109/189 (57%), Positives = 134/189 (70%)
Frame = +2
Query: 397 YNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVT 456
Y C + YQD VH+ RQF+ +C + GTVDF GN+A V QNC+++AR+PN Q+N+ T
Sbjct: 2 YKCXFVXYQDXXXVHSXRQFYRDCDVYGTVDFXXGNAATVLQNCNLYARKPNENQRNLFT 181
Query: 457 AQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDP 516
AQGR DPNQNTGI I C++ A DL VK F YLGRPWK+YSRTV++ S + D+IDP
Sbjct: 182 AQGREDPNQNTGISILNCKVAAAADLIPVKSQFKNYLGRPWKKYSRTVYLNSYMEDLIDP 361
Query: 517 VGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSS 576
GW EWNG FAL+TL Y EY N GPG+ TS RVTW G++VI +A EA FT NFI G+
Sbjct: 362 KGWLEWNGTFALDTLYYGEYNNRGPGSNTSARVTWPGYRVIKNATEANQFTVRNFIQGNE 541
Query: 577 WLGSTGFPF 585
WL ST PF
Sbjct: 542 WLSSTDIPF 568
>CK606276
Length = 751
Score = 241 bits (614), Expect = 9e-64
Identities = 117/205 (57%), Positives = 141/205 (68%), Gaps = 3/205 (1%)
Frame = +1
Query: 388 RVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRP 447
R GAD+S FYNC YQDTLYVH+ RQF+ +C I GTVDFIFGN+A + Q+C+++ R P
Sbjct: 1 RNGADMSTFYNCSFEGYQDTLYVHSLRQFYKSCDIYGTVDFIFGNAAALLQDCNMYPRLP 180
Query: 448 NSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNF---PTYLGRPWKEYSRTV 504
Q N +TAQGR DPNQNTGI IQ C I A DL N+ TYLGRPWKEYSRTV
Sbjct: 181 MQNQFNAITAQGRTDPNQNTGISIQNCCIIAASDLGDATNNYNGIKTYLGRPWKEYSRTV 360
Query: 505 FMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQ 564
+MQS I +IDP GW+EW+G+FAL+TL Y E+ N GPG+ TS RV W+G+ +I +A
Sbjct: 361 YMQSFIDGLIDPKGWNEWSGDFALSTLYYAEFANWGPGSNTSNRVAWEGYHLI-DEKDAD 537
Query: 565 SFTPGNFIGGSSWLGSTGFPFSLGL 589
FT FI G WL TG PF GL
Sbjct: 538 DFTVHKFIQGEKWLPQTGVPFXAGL 612
>TC220076 homologue to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precursor
(Pectin methylesterase 3) (PE 3) , partial (39%)
Length = 808
Score = 236 bits (602), Expect = 2e-62
Identities = 120/240 (50%), Positives = 153/240 (63%), Gaps = 1/240 (0%)
Frame = +2
Query: 347 VVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQD 406
V+ GS F +A + G F+A+DI F N AG +KHQAVA R G+D S F+ C +QD
Sbjct: 50 VLTGSMLFFAA---VKGKGFIAKDIGFVNNAGASKHQAVAFRSGSDRSVFFRCSFNGFQD 220
Query: 407 TLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQN 466
TLY H+NRQF+ +C I+GT+DFIFGN+A VFQNC I R+P Q N +TAQG+ D NQN
Sbjct: 221 TLYAHSNRQFYRDCDITGTIDFIFGNAAAVFQNCKIMPRQPLPNQFNTITAQGKKDRNQN 400
Query: 467 TGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNF 526
TGI+IQK + ++ PTYLGRPWK++S TV MQS I + PVGW W N
Sbjct: 401 TGIIIQKSKFTPLEN----NLTAPTYLGRPWKDFSTTVIMQSDIGSFLKPVGWMSWVPNV 568
Query: 527 -ALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPF 585
++T+ Y EYQNTGPGA S+RV W G+K + EA FT +FI G WL + F
Sbjct: 569 EPVSTIFYAEYQNTGPGADVSQRVKWAGYKPTLTDGEAGKFTVQSFIQGPEWLPNAAVQF 748
>TC231526 similar to UP|PME_MEDSA (Q42920) Pectinesterase precursor (Pectin
methylesterase) (PE) (P65) , partial (56%)
Length = 966
Score = 220 bits (560), Expect = 2e-57
Identities = 112/254 (44%), Positives = 157/254 (61%), Gaps = 6/254 (2%)
Frame = +2
Query: 341 ITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCD 400
ITGS+N VDG T+++AT + NF+A++I F+NTAG KHQAVALRV AD + FYNC+
Sbjct: 8 ITGSKNYVDGVQTYNTATFGVNAANFMAKNIGFENTAGAEKHQAVALRVTADKAVFYNCN 187
Query: 401 IIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGR 460
+ +QDTLY + RQF+ +C ++GT+DF+FG++ VFQNC R P Q+ +VTA GR
Sbjct: 188 MDGFQDTLYTQSQRQFYRDCTVTGTIDFVFGDAVAVFQNCKFIVRMPLENQQCLVTAGGR 367
Query: 461 VDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWH 520
+ + +V Q C ++ + YLGRPW+ Y++ V M S I D+ P G+
Sbjct: 368 SKIDSPSALVFQSCVFTGEPNVLALTPKI-AYLGRPWRLYAKVVIMDSQIDDIFVPEGYM 544
Query: 521 EWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNF--IGGS--- 575
W G+ +T Y E+ N GPGA T R+TW GFKV+ + EA + PG F I S
Sbjct: 545 AWMGSAFKDTSTYYEFNNRGPGANTIGRITWPGFKVL-NPIEAVEYYPGKFFQIANSTER 721
Query: 576 -SWLGSTGFPFSLG 588
SW+ +G P+SLG
Sbjct: 722 DSWILGSGVPYSLG 763
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.131 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,407,251
Number of Sequences: 63676
Number of extensions: 323503
Number of successful extensions: 2113
Number of sequences better than 10.0: 138
Number of HSP's better than 10.0 without gapping: 2005
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2023
length of query: 589
length of database: 12,639,632
effective HSP length: 103
effective length of query: 486
effective length of database: 6,081,004
effective search space: 2955367944
effective search space used: 2955367944
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC137821.5


