

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137821.2 + phase: 0 /pseudo
(428 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
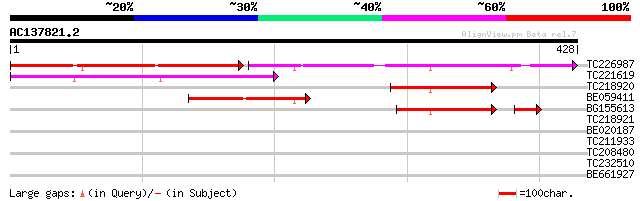
Score E
Sequences producing significant alignments: (bits) Value
TC226987 similar to UP|Q9FEV9 (Q9FEV9) Microtubule-associated pr... 166 1e-73
TC221619 similar to GB|AAP37732.1|30725420|BT008373 At1g14690 {A... 111 7e-25
TC218920 similar to GB|AAP37732.1|30725420|BT008373 At1g14690 {A... 66 3e-11
BE059411 similar to GP|20805034|db cytokinesis regulating protei... 63 2e-10
BG155613 57 4e-10
TC218921 weakly similar to GB|AAP37732.1|30725420|BT008373 At1g1... 35 0.085
BE020187 33 0.32
TC211933 similar to UP|Q9LNC4 (Q9LNC4) F9P14.9 protein, partial ... 31 1.2
TC208480 30 2.7
TC232510 homologue to UP|Q9LJQ2 (Q9LJQ2) Arabidopsis thaliana ge... 29 4.6
BE661927 28 7.9
>TC226987 similar to UP|Q9FEV9 (Q9FEV9) Microtubule-associated protein
MAP65-1a, partial (97%)
Length = 2204
Score = 166 bits (419), Expect(2) = 1e-73
Identities = 113/278 (40%), Positives = 159/278 (56%), Gaps = 30/278 (10%)
Frame = +1
Query: 181 QSISDETLARLNESVLLLKQEKQQSLQKVQFLE----ELWDFMGITTDEQKAVCDDVIGL 236
+SIS++TLARL ++VL LK++K+Q L K+Q L +LW+ M T E++ + D V
Sbjct: 898 KSISNDTLARLAKTVLTLKEDKKQRLHKLQELASQLIDLWNLMD-THPEERRLFDHVTCN 1074
Query: 237 ISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEEIH 296
+SASVDEV++ G+L+ D+++Q +VEV+RL L S+MKE +AFK+Q ELEEI
Sbjct: 1075 MSASVDEVTVPGALALDLIEQAEVEVERLDQLKASRMKE--------IAFKKQAELEEIF 1230
Query: 297 RGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAEWKS 354
AH+++D AAR+ + LI GNI+ +ELL MD QI KAKE+A SR+DI+D+V +W S
Sbjct: 1231 ARAHIEVDPDAAREKIMALIDSGNIEPTELLADMDNQIAKAKEEALSRKDILDKVEKWMS 1410
Query: 355 AAEEEKWLDEYERFI*SVRRKRG-----------------------FLSPRFHLW*RI*L 391
A EEE WL++Y R RG L + W
Sbjct: 1411 ACEEESWLEDYNRDENRYNASRGAHINLKRAEKARILVNKIPALVDTLVAKTRAW----- 1575
Query: 392 QK*KHGRQ-TKKKVSYMKSLHEYNVQRQLREEEKRKSR 428
+ HG T V + L EY + R REEEKR+ R
Sbjct: 1576 -EEDHGMSFTYDGVPLLAMLDEYAMLRHEREEEKRRMR 1686
Score = 129 bits (324), Expect(2) = 1e-73
Identities = 78/180 (43%), Positives = 112/180 (61%), Gaps = 4/180 (2%)
Frame = +3
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFS----RGK 56
MLLQ+EQE ++Y RKVE K + L + L D + E++ + S+LGE SF+
Sbjct: 354 MLLQLEQECLDVYKRKVEQAAKSRAQLLQALSDAKLELSTLLSALGEK--SFAGIPENTS 527
Query: 57 GTLKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCD 116
GT+K+QLA I PV+E L +K+ER+KE +++SQI QI E AG V+ VD+ D
Sbjct: 528 GTIKEQLAAIAPVLEQLWQQKEERIKEFSDVQSQIQQICGEIAGNLNLNDVSPA-VDESD 704
Query: 117 LTMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSS 176
L+++KL + +S L ELQ EK+ R KV +ST+ +L AV+ +D +HPSLNDSS
Sbjct: 705 LSLKKLDEYQSELQELQKEKSERLHKVLEFVSTVHDLCAVLGMDFFSTATEVHPSLNDSS 884
>TC221619 similar to GB|AAP37732.1|30725420|BT008373 At1g14690 {Arabidopsis
thaliana;} , partial (33%)
Length = 1044
Score = 111 bits (277), Expect = 7e-25
Identities = 64/209 (30%), Positives = 115/209 (54%), Gaps = 6/209 (2%)
Frame = +2
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGE----DYVSFSRGK 56
ML+++E+E E+Y RKV++ K ++ + ++E+A + ++LGE + +
Sbjct: 215 MLMELERECLEVYRRKVDEAANTKARFHQTVAAKEAELATLMAALGEHDIHSPIKTEKRS 394
Query: 57 GTLKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDV--DQ 114
+LKQ+LA+I P VE+L+ KK ER+K+ ++K+QI +I E G D
Sbjct: 395 ASLKQKLASITPWVEELKKKKDERLKQFEDVKAQIEKISGEIFGFHSVNNALSSTTVEDD 574
Query: 115 CDLTMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLND 174
DL++ +L + ++HL LQ EK+ R QKV ++ + L +V+ +D + + +HPSL+
Sbjct: 575 QDLSLRRLNEYQTHLRTLQKEKSDRLQKVLQCVNEVHSLCSVLGLDFGQTVGDVHPSLHG 754
Query: 175 SSNGAQQSISDETLARLNESVLLLKQEKQ 203
+ +IS+ TL R +K K+
Sbjct: 755 TQVEQSTNISNSTLGRSRAGHSQVKNRKE 841
>TC218920 similar to GB|AAP37732.1|30725420|BT008373 At1g14690 {Arabidopsis
thaliana;} , partial (39%)
Length = 804
Score = 65.9 bits (159), Expect = 3e-11
Identities = 34/82 (41%), Positives = 52/82 (62%), Gaps = 2/82 (2%)
Frame = +2
Query: 288 RQNELEEIHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDI 345
+++ELEEI + H + + LI G +D SELL ++ QI KAK++A SR+++
Sbjct: 2 KRSELEEICKLTHTEPYTSTTADKASALIDSGLVDPSELLANIEAQIIKAKDEALSRKEV 181
Query: 346 IDRVAEWKSAAEEEKWLDEYER 367
DR+ +W +A EEE WLDEY +
Sbjct: 182 TDRIDKWFAACEEENWLDEYNQ 247
>BE059411 similar to GP|20805034|db cytokinesis regulating protein - like
{Oryza sativa (japonica cultivar-group)}, partial (16%)
Length = 314
Score = 63.2 bits (152), Expect = 2e-10
Identities = 34/96 (35%), Positives = 59/96 (61%), Gaps = 4/96 (4%)
Frame = +1
Query: 136 KNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNGAQQSISDETLARLNESV 195
K+ R ++V+ H+ T++ L V+ D + +NGIHPSL DS +S+S++T+ +L ++
Sbjct: 1 KSSRLKQVQEHLCTLNSLCLVLGFDFKQTINGIHPSLVDSK--GSKSVSNDTIQQLAVAI 174
Query: 196 LLLKQEKQQSLQKVQFLE----ELWDFMGITTDEQK 227
L++ K Q +QK+Q L ELW+ M +EQ+
Sbjct: 175 QELREVKLQRMQKLQDLATTMLELWNLMDTPIEEQQ 282
>BG155613
Length = 381
Score = 57.0 bits (136), Expect(2) = 4e-10
Identities = 28/77 (36%), Positives = 49/77 (63%), Gaps = 2/77 (2%)
Frame = +2
Query: 293 EEIHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVA 350
EEI R H+ + A + D I G++D + +L+ ++ QI + KE+A R++I+++V
Sbjct: 2 EEICRKTHLIPEIDNAVESAVDAIESGSVDPACILEQIELQISQVKEEAFGRKEILEKVE 181
Query: 351 EWKSAAEEEKWLDEYER 367
+W +A +EE WL+EY R
Sbjct: 182 KWLAACDEESWLEEYNR 232
Score = 25.0 bits (53), Expect(2) = 4e-10
Identities = 11/20 (55%), Positives = 14/20 (70%)
Frame = +3
Query: 382 RFHLW*RI*LQK*KHGRQTK 401
+F W* +*LQK HGR+ K
Sbjct: 312 KFQQW*MV*LQKLLHGRRKK 371
>TC218921 weakly similar to GB|AAP37732.1|30725420|BT008373 At1g14690
{Arabidopsis thaliana;} , partial (9%)
Length = 276
Score = 34.7 bits (78), Expect = 0.085
Identities = 22/60 (36%), Positives = 32/60 (52%)
Frame = +1
Query: 257 QVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEEIHRGAHMDMDAKAARQILTDLI 316
Q EV RL L S+MKE V FK++++LEEI + H++ D A + + LI
Sbjct: 97 QAFAEVDRLAKLKASRMKELV--------FKKRSKLEEICKLTHIEPDTSTAAEKASALI 252
>BE020187
Length = 384
Score = 32.7 bits (73), Expect = 0.32
Identities = 20/106 (18%), Positives = 51/106 (47%)
Frame = +2
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLT 118
L++QL N +L+ K K +++ + S + ++ G+ + + ++
Sbjct: 20 LQEQLDNKCSENRELQEKVKLLEQQLATVTGGTSLMLTDQCPSGEHIDELKRKIQSQEIE 199
Query: 119 MEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEI 164
EKL + HL+E + ++++QK+ S EL++ ++++ +
Sbjct: 200 NEKLKLEQVHLSEENSGLHVQNQKLSEEASYAKELASAAAVELKNL 337
>TC211933 similar to UP|Q9LNC4 (Q9LNC4) F9P14.9 protein, partial (15%)
Length = 710
Score = 30.8 bits (68), Expect = 1.2
Identities = 25/100 (25%), Positives = 48/100 (48%), Gaps = 1/100 (1%)
Frame = +2
Query: 110 QDVDQCDLTMEKLGKLKSHLNELQNEK-NIRHQKVKSHISTISELSAVMSIDISEILNGI 168
+D + D+T E+ KL +HL L +EK + Q +K S +S+ + +DI +
Sbjct: 281 KDPHKRDMTYEEKQKLSTHLQSLPSEKLDAIVQIIKKRNSALSQHDDEIEVDIDSVDTET 460
Query: 169 HPSLNDSSNGAQQSISDETLARLNESVLLLKQEKQQSLQK 208
L+ ++S+S R E +L ++ +Q+ Q+
Sbjct: 461 LWELDRFVTNYKKSLSKN--KRKAELAILARERAEQNAQQ 574
>TC208480
Length = 1576
Score = 29.6 bits (65), Expect = 2.7
Identities = 24/91 (26%), Positives = 39/91 (42%), Gaps = 6/91 (6%)
Frame = +3
Query: 131 ELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNGAQQSISDETLAR 190
E+ +E+ IR K K + E + +I + N +H S + + +E
Sbjct: 18 EIASEETIRKLKEKHDMDVQKEAISEDTIRNLKEKNDVHVQKETLSQETIKKLKEENEVH 197
Query: 191 LNESVLL------LKQEKQQSLQKVQFLEEL 215
+ E L LK+E + LQKV LEE+
Sbjct: 198 IQEDFLSKETIKNLKEENDKLLQKVVSLEEV 290
>TC232510 homologue to UP|Q9LJQ2 (Q9LJQ2) Arabidopsis thaliana genomic DNA,
chromosome 3, P1 clone: MIE15, partial (6%)
Length = 849
Score = 28.9 bits (63), Expect = 4.6
Identities = 40/192 (20%), Positives = 78/192 (39%), Gaps = 5/192 (2%)
Frame = +3
Query: 162 SEILNGIHPSLNDSSNGAQQSISDETLARLNESVLLLKQEKQQSLQK-VQFLEELWDFMG 220
SEI++ +P ND + + + L +L E LLLK+ Q+ + V + D
Sbjct: 111 SEIVSDEYPPANDPKSNSNINDEAAELVKLTEQSLLLKKRTNQTKPRPVVVRLDDGDVAP 290
Query: 221 ITTDEQKAVCDDVIGLIS----ASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEF 276
IT + + D + G I S S+ GS +D + E ++L S+MK+
Sbjct: 291 ITVKRPEPLDDSLSGAIKDALLGSETRPSMSGSSPSDKSSR-KKEKKKLSTRVRSEMKKN 467
Query: 277 VFKRQKELAFKRQNELEEIHRGAHMDMDAKAARQILTDLIGNIDMSELLQGMDKQIRKAK 336
V + + N + H +H ++ + + + + G RK
Sbjct: 468 VVDAENP-ELENPNSSSKNHGHSHTKERRHQGKEKIVEGEEHDQREKKKSGHRHGRRKTH 644
Query: 337 EQAQSRRDIIDR 348
++A+S +++ +
Sbjct: 645 QRAKSPLNVVSQ 680
>BE661927
Length = 768
Score = 28.1 bits (61), Expect = 7.9
Identities = 16/55 (29%), Positives = 32/55 (58%)
Frame = +3
Query: 130 NELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNGAQQSIS 184
++ N++ + + K + S+ S LS +S+ I+ +LN +HPS SS A + ++
Sbjct: 12 HQSSNQQTLSNFKNQRTPSSSSSLSLSLSLVIAAVLNQVHPSSFFSSRLALKFVA 176
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.135 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,358,960
Number of Sequences: 63676
Number of extensions: 138534
Number of successful extensions: 718
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 708
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 711
length of query: 428
length of database: 12,639,632
effective HSP length: 100
effective length of query: 328
effective length of database: 6,272,032
effective search space: 2057226496
effective search space used: 2057226496
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137821.2


