

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137702.3 - phase: 2 /pseudo
(899 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
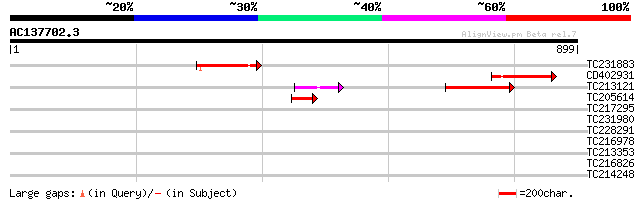
Score E
Sequences producing significant alignments: (bits) Value
TC231883 weakly similar to UP|Q7QBQ3 (Q7QBQ3) AgCP4056 (Fragment... 107 2e-23
CD402931 similar to GP|9081793|dbj P0489A01.23 {Oryza sativa (ja... 102 7e-22
TC213121 91 2e-18
TC205614 56 6e-08
TC217295 similar to UP|Q8SKU2 (Q8SKU2) Tic62 protein precursor, ... 32 0.95
TC231980 similar to UP|Q8S562 (Q8S562) KAP-2, partial (19%) 32 1.2
TC228291 homologue to UP|Q9FXT8 (Q9FXT8) 26S proteasome regulato... 31 2.1
TC216978 homologue to UP|Q8RXH5 (Q8RXH5) Osmotic stress-activate... 31 2.8
TC213353 30 3.6
TC216826 similar to UP|Q9ZPY9 (Q9ZPY9) Expressed protein (At2g46... 30 3.6
TC214248 similar to UP|Q9FMJ2 (Q9FMJ2) Ankyrin-like protein, par... 29 8.1
>TC231883 weakly similar to UP|Q7QBQ3 (Q7QBQ3) AgCP4056 (Fragment), partial
(17%)
Length = 1263
Score = 107 bits (268), Expect = 2e-23
Identities = 54/110 (49%), Positives = 74/110 (67%), Gaps = 7/110 (6%)
Frame = +1
Query: 297 LHPN-------QIDKIISAEIPDKNRDPKLYEIVASLMIHGPCGPQNKSSPCILIKKCTK 349
LHP+ +ID IISAEIP DP+LY +V + M+HGPCG SPC+ KC++
Sbjct: 934 LHPDNQYPSSDEIDHIISAEIPSHEDDPELYTLVQNHMVHGPCGILQSHSPCMKEGKCSR 1113
Query: 350 YFPKKFVDNTVIDSDGYPVYRRRDNGVFIKKKGESFVDNRWVVPYNRKLM 399
++PK F+ NT++D +GY VYRRR++G I K VDNR++VPYN KL+
Sbjct: 1114 FYPKIFLPNTLLDLNGYLVYRRRNDGRTI-LKNSVIVDNRYLVPYNAKLL 1260
>CD402931 similar to GP|9081793|dbj P0489A01.23 {Oryza sativa (japonica
cultivar-group)}, partial (5%)
Length = 617
Score = 102 bits (254), Expect = 7e-22
Identities = 51/103 (49%), Positives = 73/103 (70%)
Frame = +2
Query: 764 GTGKTFILRALSSAVRSRKEIVLTVASSGIAALLIPGGRTAHSRFGIPIIVDEISTCGIH 823
GT KTF+ + LS+ +RS K ++ + +A+LL+P G TAHS F IP+++ + STC I+
Sbjct: 11 GTSKTFLWKTLSAGMRS-KGLMSSSCLKRLASLLLPRG*TAHSTFSIPLVIKDDSTCNIN 187
Query: 824 PKSPLAKLVCKAKLIIWDEAPMMHKHCFEALDRSLRDILRVQN 866
S +KL+ KLIIWDE P+M+K CFEALDR+L+DI+ QN
Sbjct: 188 QGSARSKLLLHTKLIIWDETPVMNKFCFEALDRTLQDIMASQN 316
>TC213121
Length = 713
Score = 90.9 bits (224), Expect = 2e-18
Identities = 43/109 (39%), Positives = 70/109 (63%)
Frame = +2
Query: 692 DYPTMPRTDISLIHESRNRLIYDELNYNQQLLEIEHKKLMSTMTAEQKNVYEKIISRVDD 751
++P M +++ N+LIY+EL Y++ +L + ++T E +++ KI+ +
Sbjct: 284 NFPLMSYPIGYIVNPYGNKLIYNELAYDKDILAA*FSRCYHSLTDEHASIFNKIMHMIAT 463
Query: 752 NLPGIFFLYGYGGTGKTFILRALSSAVRSRKEIVLTVASSGIAALLIPG 800
G++FLYGYGGTGKTF+ + LSS + S IVLT+ASSGI ++L+PG
Sbjct: 464 QSGGVYFLYGYGGTGKTFVWKTLSSTIHSNSGIVLTMASSGILSMLLPG 610
Score = 48.5 bits (114), Expect = 1e-05
Identities = 27/77 (35%), Positives = 45/77 (58%)
Frame = +3
Query: 452 CRYLSACEAVWRIFSFDINYREPSVERLNFHLEGEEPVVFEDHEDIADVIKKPHIRDTKF 511
C Y+S E +I +F ++ R P+VE L FHLE + P ++ + I ++ K I+++ F
Sbjct: 30 CIYVSPPETC*KISAFPMHGRAPTVECLYFHLENQ*P--*KESQQIGSMLSKSTIKESIF 203
Query: 512 IAWFEANQKYPEARDLT 528
A ++N+ Y RDLT
Sbjct: 204 TACMDSNKMYHHGRDLT 254
>TC205614
Length = 742
Score = 56.2 bits (134), Expect = 6e-08
Identities = 27/41 (65%), Positives = 30/41 (72%), Gaps = 1/41 (2%)
Frame = +2
Query: 448 MYY-DCRYLSACEAVWRIFSFDINYREPSVERLNFHLEGEE 487
MYY DC+Y+S CEA WRIFSF I R P VERL FHL E+
Sbjct: 617 MYYLDCQYISPCEACWRIFSFQIYGRIPIVERLYFHLPNEQ 739
>TC217295 similar to UP|Q8SKU2 (Q8SKU2) Tic62 protein precursor, partial
(21%)
Length = 609
Score = 32.3 bits (72), Expect = 0.95
Identities = 15/34 (44%), Positives = 22/34 (64%)
Frame = -1
Query: 652 TWQNLSDDMLHRQRRASQLKNYTLAEIDRLLRSH 685
TW+ LS D+LH+++ AS L L+E LL +H
Sbjct: 156 TWEALSQDILHKEKEASPL*MTMLSESRHLLPAH 55
>TC231980 similar to UP|Q8S562 (Q8S562) KAP-2, partial (19%)
Length = 590
Score = 32.0 bits (71), Expect = 1.2
Identities = 12/35 (34%), Positives = 24/35 (68%)
Frame = -3
Query: 634 VQLLVTNQFAQPEVVWSKTWQNLSDDMLHRQRRAS 668
V LL+T ++ E VW +TW ++DD+++ +++S
Sbjct: 588 VLLLLTATXSKSEQVWDQTWHWMADDIVYNYKKSS 484
>TC228291 homologue to UP|Q9FXT8 (Q9FXT8) 26S proteasome regulatory particle
triple-A ATPase subunit4, partial (62%)
Length = 1079
Score = 31.2 bits (69), Expect = 2.1
Identities = 25/85 (29%), Positives = 37/85 (43%), Gaps = 7/85 (8%)
Frame = +3
Query: 744 KIISRVDDNLPGIFFLYGYGGTGKTFILRALSSAVRSRKEIVLTVASSGIAALLIPGGRT 803
++ RV P LYG GTGKT + RA++S + + + V+S+ I + R
Sbjct: 48 ELFIRVGIKPPKGVLLYGPPGTGKTLLARAIASNIEA--NFLKVVSSAIIDKYIGESARL 221
Query: 804 AHSRFGIP-------IIVDEISTCG 821
FG I +DEI G
Sbjct: 222 IREMFGYARDHQPCIIFMDEIDAIG 296
>TC216978 homologue to UP|Q8RXH5 (Q8RXH5) Osmotic stress-activated protein
kinase, partial (96%)
Length = 1626
Score = 30.8 bits (68), Expect = 2.8
Identities = 14/31 (45%), Positives = 18/31 (57%)
Frame = -1
Query: 75 ITMPPTSKAYFRVRNILFAIAIPFNLFKGRR 105
I +P TS Y RNIL + PFN+F G +
Sbjct: 531 IRVPKTSMVYDFPRNILINLVTPFNVFHGNK 439
>TC213353
Length = 392
Score = 30.4 bits (67), Expect = 3.6
Identities = 17/35 (48%), Positives = 22/35 (62%)
Frame = -1
Query: 758 FLYGYGGTGKTFILRALSSAVRSRKEIVLTVASSG 792
+L YG GK+ ILR +S RSR+E V + A SG
Sbjct: 263 YLVSYGSQGKSSILR--TSRSRSRRESVTSAAGSG 165
>TC216826 similar to UP|Q9ZPY9 (Q9ZPY9) Expressed protein
(At2g46500/F11C10.19), partial (39%)
Length = 850
Score = 30.4 bits (67), Expect = 3.6
Identities = 21/62 (33%), Positives = 33/62 (52%)
Frame = -2
Query: 769 FILRALSSAVRSRKEIVLTVASSGIAALLIPGGRTAHSRFGIPIIVDEISTCGIHPKSPL 828
F RAL+ + R RK ++ ASS +A L+PG + + I I D + +H + P+
Sbjct: 747 FYKRALTISSR-RKSMIWETASSNVA-SLVPGRTESSAACTISSITDSLFKDSLHIRLPM 574
Query: 829 AK 830
AK
Sbjct: 573 AK 568
>TC214248 similar to UP|Q9FMJ2 (Q9FMJ2) Ankyrin-like protein, partial (60%)
Length = 1596
Score = 29.3 bits (64), Expect = 8.1
Identities = 17/41 (41%), Positives = 22/41 (53%), Gaps = 5/41 (12%)
Frame = +3
Query: 657 SDDMLHRQRRASQLKNY-----TLAEIDRLLRSHGKSMKED 692
S D+LH S +KN +AEIDR+LR GK + D
Sbjct: 1131 SYDLLHADNLFSNIKNRCNLKAVVAEIDRILRPEGKLIVRD 1253
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.335 0.146 0.476
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,503,039
Number of Sequences: 63676
Number of extensions: 647275
Number of successful extensions: 4651
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 4610
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4649
length of query: 899
length of database: 12,639,632
effective HSP length: 106
effective length of query: 793
effective length of database: 5,889,976
effective search space: 4670750968
effective search space used: 4670750968
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC137702.3


