

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137670.4 + phase: 0
(324 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
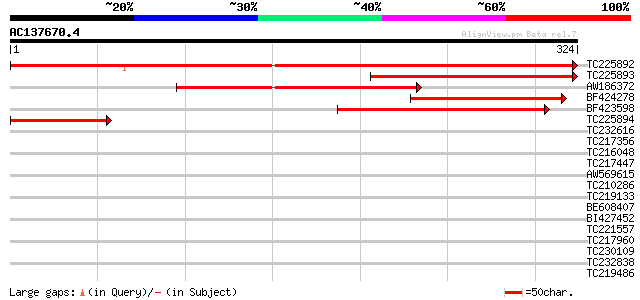
Score E
Sequences producing significant alignments: (bits) Value
TC225892 similar to UP|ARG3_HUMAN (Q9NP61) ADP-ribosylation fact... 487 e-138
TC225893 similar to UP|Q9FIQ0 (Q9FIQ0) Zinc finger protein Glo3-... 189 2e-48
AW186372 172 2e-43
BF424278 142 2e-34
BF423598 weakly similar to GP|9758511|dbj| zinc finger protein G... 135 2e-32
TC225894 similar to UP|Q9FIQ0 (Q9FIQ0) Zinc finger protein Glo3-... 106 1e-23
TC232616 similar to GB|AAD25756.1|4587525|T5I8 Contains the PF|0... 36 0.027
TC217356 similar to UP|Q84JM5 (Q84JM5) Angustifolia, partial (57%) 35 0.035
TC216048 similar to UP|Q8S490 (Q8S490) Transcription factor RAU1... 35 0.060
TC217447 similar to GB|AAM65362.1|21655301|AY103310 At2g37550/F1... 33 0.18
AW569615 32 0.30
TC210286 similar to UP|Q9FH69 (Q9FH69) Arabidopsis thaliana geno... 31 0.67
TC219133 UP|Q39879 (Q39879) Mitotic cyclin a2-type, complete 31 0.67
BE608407 similar to GP|4519792|dbj Asp1 {Arabidopsis thaliana}, ... 31 0.87
BI427452 31 0.87
TC221557 similar to UP|Q43614 (Q43614) DNA-binding protein, part... 31 0.87
TC217960 weakly similar to GB|AAF48440.1|7293054|AE003498 CG6146... 30 1.1
TC230109 homologue to UP|Q8LDR0 (Q8LDR0) Zinc finger protein OBP... 30 1.1
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 30 1.5
TC219486 29 3.3
>TC225892 similar to UP|ARG3_HUMAN (Q9NP61) ADP-ribosylation factor
GTPase-activating protein 3 (ARF GAP 3), partial (12%)
Length = 1727
Score = 487 bits (1254), Expect = e-138
Identities = 255/327 (77%), Positives = 287/327 (86%), Gaps = 3/327 (0%)
Frame = +2
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALEKGLPSSPVA 60
MSFGGN+RA FFKQHGWTDGGKIEAKYTSRAA+LYRQIL+KEVAKSMA + GLPSSPVA
Sbjct: 440 MSFGGNNRAHGFFKQHGWTDGGKIEAKYTSRAADLYRQILSKEVAKSMAEDGGLPSSPVA 619
Query: 61 SQSS---NGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKPGKSG 117
SQS+ NG +V+T+EV KENTL+K EK ESTSSPRASH+ S +KK IG KK KSG
Sbjct: 620 SQSAQGVNGLPEVKTNEVPKENTLEKPEKPESTSSPRASHSVISGTVKKPIGAKKAVKSG 799
Query: 118 GLGARKLNKKPSESFYEQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSRG 177
GLGARKL KKPSES YEQKPEEPPAPVPS+TNN + A PS TSRFEYV+NVQSS+L++ G
Sbjct: 800 GLGARKLTKKPSESLYEQKPEEPPAPVPSSTNN-MPAGPSPTSRFEYVENVQSSDLNTGG 976
Query: 178 SNTFNHVSVPKSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNAKSISSSQFF 237
S+ +HVS PKSS+FFADFGMD GFPKK G ++SKVQI+E+DEAR+KFSNAKSISSSQFF
Sbjct: 977 SHVLSHVSPPKSSSFFADFGMDGGFPKKSGPSSSKVQIQETDEARRKFSNAKSISSSQFF 1156
Query: 238 GDQNKARDAETRATLSKFSSSSAISSADFFGDSADSSIDLAASDLINRLSFQAQQDISSL 297
GDQNKA D +++ATLSKFS SSAISSAD FGDS D++IDL A DLINRLSFQAQQD+SSL
Sbjct: 1157GDQNKAADVDSQATLSKFSGSSAISSADLFGDSRDNNIDLTAGDLINRLSFQAQQDLSSL 1336
Query: 298 KNIAGETGKKLSSLASSLMTDLQDRIL 324
KNIAGETGKKLSSLAS+LMTDLQDRIL
Sbjct: 1337KNIAGETGKKLSSLASTLMTDLQDRIL 1417
>TC225893 similar to UP|Q9FIQ0 (Q9FIQ0) Zinc finger protein Glo3-like,
partial (28%)
Length = 674
Score = 189 bits (479), Expect = 2e-48
Identities = 97/118 (82%), Positives = 110/118 (93%)
Frame = +1
Query: 207 GSNTSKVQIEESDEARKKFSNAKSISSSQFFGDQNKARDAETRATLSKFSSSSAISSADF 266
G ++SKVQI+E+DEAR+KFSNAKSISSSQ+FGDQNKA D +++ATLSKFS SSAISSAD
Sbjct: 1 GPSSSKVQIQETDEARRKFSNAKSISSSQYFGDQNKAADVDSQATLSKFSGSSAISSADL 180
Query: 267 FGDSADSSIDLAASDLINRLSFQAQQDISSLKNIAGETGKKLSSLASSLMTDLQDRIL 324
FGDS D++IDL A DLINRLSFQAQQD+SSLKNIAGETGKKLSSLAS+LMTDLQDRIL
Sbjct: 181 FGDSRDNNIDLTAGDLINRLSFQAQQDLSSLKNIAGETGKKLSSLASTLMTDLQDRIL 354
>AW186372
Length = 418
Score = 172 bits (435), Expect = 2e-43
Identities = 91/140 (65%), Positives = 107/140 (76%)
Frame = +2
Query: 96 SHTSASNNLKKSIGGKKPGKSGGLGARKLNKKPSESFYEQKPEEPPAPVPSTTNNNVSAR 155
S + S K IG KK KSGGLGARKL KKPSES YEQKPEEPPAPVPS+TN+ + A
Sbjct: 2 SDSVISGTGNKPIGAKKAVKSGGLGARKLTKKPSESLYEQKPEEPPAPVPSSTNS-MPAG 178
Query: 156 PSMTSRFEYVDNVQSSELDSRGSNTFNHVSVPKSSNFFADFGMDSGFPKKFGSNTSKVQI 215
PS TSRFEYV+NVQS +L++ GS+ +HV PKSS+FFADFGMD G PKK G ++ KVQI
Sbjct: 179 PSPTSRFEYVENVQSCDLNTGGSHVLSHVYPPKSSSFFADFGMDGGCPKKSGPSSCKVQI 358
Query: 216 EESDEARKKFSNAKSISSSQ 235
E+DEAR K NAK+ SSS+
Sbjct: 359 HETDEARMKCVNAKADSSSE 418
>BF424278
Length = 417
Score = 142 bits (359), Expect = 2e-34
Identities = 73/89 (82%), Positives = 82/89 (92%)
Frame = +3
Query: 230 SISSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFGDSADSSIDLAASDLINRLSFQ 289
+ISSSQ+FGDQNKA D +++ATLSKFS SSAISSAD FGDS D++IDL A DLINRLSFQ
Sbjct: 150 AISSSQYFGDQNKAADVDSQATLSKFSGSSAISSADLFGDSRDNNIDLTAGDLINRLSFQ 329
Query: 290 AQQDISSLKNIAGETGKKLSSLASSLMTD 318
AQQD+SSLKNIAGETGKKLSSLAS+LMTD
Sbjct: 330 AQQDLSSLKNIAGETGKKLSSLASTLMTD 416
>BF423598 weakly similar to GP|9758511|dbj| zinc finger protein Glo3-like
{Arabidopsis thaliana}, partial (20%)
Length = 364
Score = 135 bits (341), Expect = 2e-32
Identities = 71/121 (58%), Positives = 91/121 (74%)
Frame = +2
Query: 188 KSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNAKSISSSQFFGDQNKARDAE 247
K S+FFAD GMD GF KK G ++SKVQI+E+DE+R+KFS+AKSISSSQ GD N+A D +
Sbjct: 2 K*SSFFADLGMDGGFAKKSGPSSSKVQIQETDESRRKFSHAKSISSSQILGDHNRAADVD 181
Query: 248 TRATLSKFSSSSAISSADFFGDSADSSIDLAASDLINRLSFQAQQDISSLKNIAGETGKK 307
ATL+K+++ SAISSAD G D+++DL A DL +R S QA QD+S LKNI+ ET
Sbjct: 182 DEATLTKWTAPSAISSADLVGC*RDNTVDLTAGDLSHRSSMQAPQDLSVLKNISRETRST 361
Query: 308 L 308
L
Sbjct: 362 L 364
>TC225894 similar to UP|Q9FIQ0 (Q9FIQ0) Zinc finger protein Glo3-like,
partial (32%)
Length = 612
Score = 106 bits (265), Expect = 1e-23
Identities = 50/58 (86%), Positives = 55/58 (94%)
Frame = +1
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALEKGLPSSP 58
MSFGGN+RAQ+FFKQHGW DGGKIEAKYTSRAA+LYRQIL+KEVAKSMA + GLPSSP
Sbjct: 439 MSFGGNNRAQVFFKQHGWNDGGKIEAKYTSRAADLYRQILSKEVAKSMAEDGGLPSSP 612
>TC232616 similar to GB|AAD25756.1|4587525|T5I8 Contains the PF|00650
CRAL/TRIO phosphatidyl-inositol-transfer protein domain.
ESTs gb|T76582, gb|N06574 and gb|Z25700 come from this
gene. {Arabidopsis thaliana;} , partial (48%)
Length = 1214
Score = 35.8 bits (81), Expect = 0.027
Identities = 38/141 (26%), Positives = 57/141 (39%), Gaps = 1/141 (0%)
Frame = -3
Query: 185 SVPKSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNAKSISSSQFFGDQNKAR 244
S PKSS+ DS + + S + + A K F+ S S+ G + +
Sbjct: 645 SEPKSSSTTESI-FDSFLHLRVFFSISNASLTLNSLALKNFNRTTSTPSAPLLGKRVTPQ 469
Query: 245 -DAETRATLSKFSSSSAISSADFFGDSADSSIDLAASDLINRLSFQAQQDISSLKNIAGE 303
D +T + FSSSS+ SS FF S S + + D+SSLK
Sbjct: 468 IDTSFSSTSTLFSSSSSFSSPSFFFSSFSGKASFFGSKRVFPKIASSSFDLSSLKAFLSN 289
Query: 304 TGKKLSSLASSLMTDLQDRIL 324
+ + L SSL +L +L
Sbjct: 288 SLRSER*LLSSL*EELFSTVL 226
>TC217356 similar to UP|Q84JM5 (Q84JM5) Angustifolia, partial (57%)
Length = 1672
Score = 35.4 bits (80), Expect = 0.035
Identities = 27/99 (27%), Positives = 42/99 (42%)
Frame = +3
Query: 82 DKAEKLESTSSPRASHTSASNNLKKSIGGKKPGKSGGLGARKLNKKPSESFYEQKPEEPP 141
D +K ST +S ++L +S + G+ G +K K+ + +QKPE+P
Sbjct: 633 DNFQKKVSTQMKESSSQHQVSSLSQSTSARSEGRRSRSG-KKAKKRHTRQKSQQKPEDPS 809
Query: 142 APVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSRGSNT 180
AP T+ S T + + SS DSR T
Sbjct: 810 APEKEGTSRRDDTAMSGTDQ-----ALSSSSEDSRSRKT 911
>TC216048 similar to UP|Q8S490 (Q8S490) Transcription factor RAU1 (Fragment),
partial (73%)
Length = 1933
Score = 34.7 bits (78), Expect = 0.060
Identities = 42/221 (19%), Positives = 85/221 (38%), Gaps = 5/221 (2%)
Frame = +2
Query: 27 KYTSRAAELYRQILTKEVAKSMALEKGLPSSPVASQSSNGFLDVRTSEVLKENTLDKAE- 85
+Y S + + I+ +E + + PSSP + + F++ SE +E++L +
Sbjct: 425 RYRSAPSSYFSNIIDREFYEHVF---NRPSSPETERVFSRFMNSLNSE--EEDSLHHHKL 589
Query: 86 KLESTSSPRASHTSASNNLKKSIGGKKPG----KSGGLGARKLNKKPSESFYEQKPEEPP 141
+S+SS A N +S+ + + N S +FY+ +PP
Sbjct: 590 STDSSSSSAAVKEEVVNQHNQSVNEEHVVVAALQQSNNNMNSYNNSASRNFYQSSSSKPP 769
Query: 142 APVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSRGSNTFNHVSVPKSSNFFADFGMDSG 201
P P+ P+++S E SN H S P + F+ +++
Sbjct: 770 LPNPN---------PNLSSGMEQGSFSMGLRHSGNNSNLIRHSSSP--AGLFSQINIENV 916
Query: 202 FPKKFGSNTSKVQIEESDEARKKFSNAKSISSSQFFGDQNK 242
+ G T ++A KFS+++ + + + +
Sbjct: 917 YAGVRGMGTLGAVNNSIEDA--KFSSSRRLKNQPNYSSSGR 1033
>TC217447 similar to GB|AAM65362.1|21655301|AY103310 At2g37550/F13M22.5
{Arabidopsis thaliana;} , partial (35%)
Length = 563
Score = 33.1 bits (74), Expect = 0.18
Identities = 24/81 (29%), Positives = 33/81 (40%), Gaps = 14/81 (17%)
Frame = +3
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQIL--------------TKEVAK 46
M GGN + F Q+G I AKY S AA +YR + KE +
Sbjct: 252 MEAGGNDKLNAFLTQYGIPKETDIVAKYNSNAAAVYRDRIQALADGRPWRDPPVVKEAVR 431
Query: 47 SMALEKGLPSSPVASQSSNGF 67
S A KG P + ++ G+
Sbjct: 432 SSA-SKGKPPLSAGNNNNGGW 491
>AW569615
Length = 421
Score = 32.3 bits (72), Expect = 0.30
Identities = 18/43 (41%), Positives = 24/43 (54%)
Frame = -2
Query: 77 KENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKPGKSGGL 119
K++T+ L S+S PR H S NN + + G KK G GGL
Sbjct: 354 KDHTISSHHHLPSSSVPRFHHVS--NNERATNGPKKGGSGGGL 232
>TC210286 similar to UP|Q9FH69 (Q9FH69) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K16E1, partial (8%)
Length = 560
Score = 31.2 bits (69), Expect = 0.67
Identities = 34/146 (23%), Positives = 62/146 (42%)
Frame = +3
Query: 55 PSSPVASQSSNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKPG 114
PSSP +S SS ++ + AEKL S+ P+A +T A
Sbjct: 102 PSSPTSSSSS-------------QHQKEVAEKLGSSQVPKAPYTVA-------------- 200
Query: 115 KSGGLGARKLNKKPSESFYEQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSSELD 174
GL ++K N P+ E +P + A PS+++ N + P + ++ S
Sbjct: 201 ---GLSSQKSNPIPA----EDEPMQIEAAPPSSSSANENENPPLEDTI-----MEESIRV 344
Query: 175 SRGSNTFNHVSVPKSSNFFADFGMDS 200
+RG N + + ++ F+ F +++
Sbjct: 345 TRGRLRKNRSAGIR*TSMFSSFSLEN 422
>TC219133 UP|Q39879 (Q39879) Mitotic cyclin a2-type, complete
Length = 1804
Score = 31.2 bits (69), Expect = 0.67
Identities = 31/110 (28%), Positives = 50/110 (45%), Gaps = 18/110 (16%)
Frame = +2
Query: 99 SASNNLKKSIGGKKPGKSGGLGARKLNKKPSESFYEQKPEEP------PAPV----PSTT 148
+ S N + ++ KP + + K N P ++ P P PAPV PS +
Sbjct: 272 ACSGNKRPTLANVKPASA--VIFPKANSFPVKNEAPPTPPPPVATVTVPAPVVDVSPSKS 445
Query: 149 NN-NVSARPSMTS-------RFEYVDNVQSSELDSRGSNTFNHVSVPKSS 190
+ +VS SM+S EYVDN + +DS TF+H+++ S+
Sbjct: 446 DTMSVSTDESMSSCDSFKSPDIEYVDNSDVAAVDSIERKTFSHLNISDST 595
>BE608407 similar to GP|4519792|dbj Asp1 {Arabidopsis thaliana}, partial
(26%)
Length = 421
Score = 30.8 bits (68), Expect = 0.87
Identities = 15/37 (40%), Positives = 19/37 (50%)
Frame = +2
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYR 37
M GGN + F Q+G I KY+S AA +YR
Sbjct: 212 MEAGGNDKLNAFLLQYGIPKETDIVVKYSSNAASVYR 322
>BI427452
Length = 434
Score = 30.8 bits (68), Expect = 0.87
Identities = 29/108 (26%), Positives = 44/108 (39%)
Frame = +3
Query: 55 PSSPVASQSSNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKPG 114
PSSP + SS TS TL+ + + +T SPRA+ TS+ N+ I P
Sbjct: 18 PSSPNSPSSSPKAPPPPTSFASSPKTLESSTSISTTPSPRAT-TSSKNSCFPMIPPSAPS 194
Query: 115 KSGGLGARKLNKKPSESFYEQKPEEPPAPVPSTTNNNVSARPSMTSRF 162
N P+ SF PP+ + ++ +S P + F
Sbjct: 195 ----------NHAPASSF-----RTPPSSTFNFSSTTMSRNPRLLCCF 293
>TC221557 similar to UP|Q43614 (Q43614) DNA-binding protein, partial (39%)
Length = 1114
Score = 30.8 bits (68), Expect = 0.87
Identities = 22/77 (28%), Positives = 34/77 (43%), Gaps = 8/77 (10%)
Frame = +3
Query: 80 TLDKAEKLESTSSPRASHTSASNNLK--------KSIGGKKPGKSGGLGARKLNKKPSES 131
T +K ++ A+ T+A+NN K K+ P G R +KKP +
Sbjct: 411 TEEKGNSSNVNTTTAATTTAANNNTKVGFYIYECKTCNRTFPSFQALGGHRASHKKPKLA 590
Query: 132 FYEQKPEEPPAPVPSTT 148
E+K PP+P+P T
Sbjct: 591 AEEKKQPLPPSPLPPPT 641
>TC217960 weakly similar to GB|AAF48440.1|7293054|AE003498 CG6146-PA
{Drosophila melanogaster;} , partial (3%)
Length = 1043
Score = 30.4 bits (67), Expect = 1.1
Identities = 32/100 (32%), Positives = 43/100 (43%), Gaps = 2/100 (2%)
Frame = -3
Query: 218 SDEARKKFSNAKSISSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFGDSADSSIDL 277
S + K S++KSISSS D + A T TL + +S S DS I L
Sbjct: 795 SSSSSSKSSSSKSISSSSSTSDSSAT--AATPFTLPLSNHCLPLSFFSLGAVSTDSGISL 622
Query: 278 AASDL--INRLSFQAQQDISSLKNIAGETGKKLSSLASSL 315
A+S+ + LS A KN++ TG SSL
Sbjct: 621 ASSNASSASNLSLHA-------KNVSTVTGSPWGITTSSL 523
>TC230109 homologue to UP|Q8LDR0 (Q8LDR0) Zinc finger protein OBP4-like,
partial (27%)
Length = 696
Score = 30.4 bits (67), Expect = 1.1
Identities = 21/75 (28%), Positives = 31/75 (41%)
Frame = +2
Query: 117 GGLGARKLNKKPSESFYEQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSR 176
GG K + KP++ + PP P P NN+ S S +SS L +
Sbjct: 371 GGCRKSKRSSKPNKITPSETASPPPPPHPDHNNNSNSHSSS-----------ESSSLTAA 517
Query: 177 GSNTFNHVSVPKSSN 191
+ T VS P++ N
Sbjct: 518 AATTTEAVSAPETLN 562
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 30.0 bits (66), Expect = 1.5
Identities = 30/115 (26%), Positives = 46/115 (39%), Gaps = 4/115 (3%)
Frame = -3
Query: 170 SSELDSRGSNTFN----HVSVPKSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKF 225
S LD+ S+ + SVPKSS+ S ++S+ S +
Sbjct: 636 SKSLDASSSSAVSKGRIRYSVPKSSSSPCSSSSSSSSSSSSSESSSESLSSSSGSPPRWT 457
Query: 226 SNAKSISSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFGDSADSSIDLAAS 280
S + S SSS + A + + + SSSS+ SS+ S SS D +S
Sbjct: 456 SPSSSSSSSSSSSPPSGASSSSSSLSSPSSSSSSSSSSSSSSASSTTSSTDSYSS 292
>TC219486
Length = 1900
Score = 28.9 bits (63), Expect = 3.3
Identities = 39/166 (23%), Positives = 62/166 (36%), Gaps = 3/166 (1%)
Frame = +2
Query: 127 KPSES---FYEQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSRGSNTFNH 183
+PS+S + KP PP TT R + ++ S +D +F H
Sbjct: 122 EPSDSASTLLDHKPHSPP-----TTIRRRPLRRGIPGTQSSDSSIVSDLIDVDSRRSFRH 286
Query: 184 VSVPKSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNAKSISSSQFFGDQNKA 243
S ++ N + +K S + Q+ S E + S S ++ GD
Sbjct: 287 KSRLRNLN------KNENSEEKPDSEEPR-QVNASPEENNEGSTVTSAANDDAAGD---- 433
Query: 244 RDAETRATLSKFSSSSAISSADFFGDSADSSIDLAASDLINRLSFQ 289
+I SA GDS+ S ++LAA +I+ L FQ
Sbjct: 434 ----------------SIDSAPRXGDSSSSLLELAAGLVISLLGFQ 523
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.306 0.123 0.324
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,471,786
Number of Sequences: 63676
Number of extensions: 121272
Number of successful extensions: 1013
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 958
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 997
length of query: 324
length of database: 12,639,632
effective HSP length: 97
effective length of query: 227
effective length of database: 6,463,060
effective search space: 1467114620
effective search space used: 1467114620
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC137670.4


