

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137603.6 - phase: 0 /pseudo
(1211 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
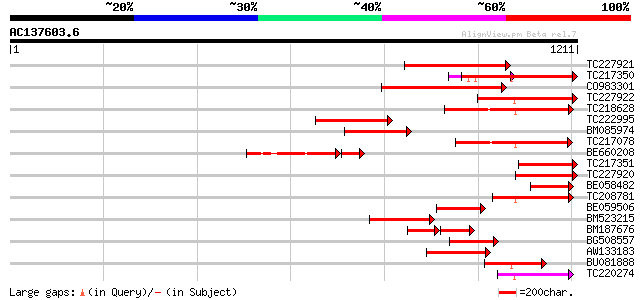
Score E
Sequences producing significant alignments: (bits) Value
TC227921 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like pro... 382 e-106
TC217350 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like pro... 382 e-106
CO983301 373 e-103
TC227922 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like pro... 292 7e-79
TC218628 similar to UP|Q9LWU7 (Q9LWU7) ESTs AU033035(S1515), par... 277 2e-74
TC222995 weakly similar to UP|GLS2_YEAST (P40989) 1,3-beta-gluca... 274 2e-73
BM085974 homologue to PIR|T49914|T4 callose synthase catalytic s... 273 3e-73
TC217078 272 7e-73
BE660208 246 3e-71
TC217351 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like pro... 209 6e-54
TC227920 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like pro... 207 3e-53
BE058482 181 2e-45
TC208781 similar to UP|Q9SHJ3 (Q9SHJ3) F12K11.17, partial (8%) 179 7e-45
BE059506 similar to PIR|F86200|F8 protein F12K11.17 [imported] -... 171 2e-42
BM523215 169 9e-42
BM187676 98 4e-38
BG508557 weakly similar to GP|9294379|dbj glucan synthase-like p... 150 3e-36
AW133183 weakly similar to PIR|T49914|T49 callose synthase catal... 137 3e-32
BU081888 129 8e-30
TC220274 116 7e-26
>TC227921 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like protein
(Fragment), partial (27%)
Length = 759
Score = 382 bits (982), Expect = e-106
Identities = 183/227 (80%), Positives = 203/227 (88%), Gaps = 2/227 (0%)
Frame = +3
Query: 844 GFLMALPMLMEIGLERGFRTALSEFILMQLQLAPVFFTFSLGTKTHYYGRTLLHGGAKYR 903
GFL+ALPMLMEIGLERGFR ALSEF+LMQLQLAPVFFTFSLGTKTHYYGRTLLHGGA+Y+
Sbjct: 3 GFLLALPMLMEIGLERGFREALSEFVLMQLQLAPVFFTFSLGTKTHYYGRTLLHGGAEYK 182
Query: 904 PTGRGFVVFHAKFADNYRLYSRSHFVKGIELMILLIVYQIFGNGYRSGLSYLLITTPMWF 963
TGRGFVVFHAKFADNYRLYSRSHFVKGIELMILL+VY IFG+ YR L+Y+LIT MWF
Sbjct: 183 STGRGFVVFHAKFADNYRLYSRSHFVKGIELMILLVVYHIFGHEYRGVLAYILITVTMWF 362
Query: 964 MVGTWLYAPFLFNPSGFEWQKIVDDWTDWNKWISIRGGIGVPPEKSWESWWEEEQEHLKY 1023
MVGTWL+APFLFNPSGFEWQKIVDD+TDW KWIS RGGIGV P+KSWESWWE+E EHL++
Sbjct: 363 MVGTWLFAPFLFNPSGFEWQKIVDDYTDWQKWISNRGGIGVSPQKSWESWWEKEHEHLRH 542
Query: 1024 SGMRGIIAEILLSLRFFIYQYGLVYHLNFT-KSTKSVLPTGL-WHIM 1068
SG R I EI+L+ RFFIYQYGLVYHL+ T + T+SVL GL W I+
Sbjct: 543 SGKRRIATEIILTRRFFIYQYGLVYHLSVTDEKTQSVLVYGLSWMII 683
>TC217350 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like protein
(Fragment), partial (31%)
Length = 1049
Score = 382 bits (981), Expect = e-106
Identities = 195/261 (74%), Positives = 214/261 (81%), Gaps = 15/261 (5%)
Frame = +2
Query: 966 GTWLYAPFLFNPS-----GFEWQKIVDDWTDWNKWISIRGGIGVPPEKSWESWWEEEQEH 1020
G W + P PS G +KIVDDWTDWNKWIS RGGIGVPPEKSWESWWEEEQEH
Sbjct: 77 GLW-WVPGCLLPSCSILLGLNGKKIVDDWTDWNKWISNRGGIGVPPEKSWESWWEEEQEH 253
Query: 1021 LKYSGMRGIIAEILLSLRFFIYQYGLVYHLNFTKS-TKSVLPTGLWHIMVG--------- 1070
L+YSGMRGII EILLSLRFFIYQYGLVYHLN TK TKS L G+ +++
Sbjct: 254 LQYSGMRGIIVEILLSLRFFIYQYGLVYHLNITKKGTKSFLVYGISWLVIFVILFVMKTV 433
Query: 1071 DLSNISYLEDFQLVFRLMKGLVFVTFVSILVTMIALAHMTLQDIVVCILAFMPTGWGMLQ 1130
+ + +FQLVFRL+KG++F+TFVSILV +IAL HMT+QDIVVCILAFMPTGWGMLQ
Sbjct: 434 SVGRRKFSANFQLVFRLIKGMIFLTFVSILVILIALPHMTVQDIVVCILAFMPTGWGMLQ 613
Query: 1131 IAQALKPLVRRGGFWESVKTLARGYEVIMGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFS 1190
IAQALKP+VRR GFW SVKTLARGYE++MGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFS
Sbjct: 614 IAQALKPVVRRAGFWGSVKTLARGYEIVMGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFS 793
Query: 1191 RGLQISRILGGQRKGRSSRNK 1211
RGLQISRILGGQRK RSSRNK
Sbjct: 794 RGLQISRILGGQRKERSSRNK 856
Score = 90.5 bits (223), Expect = 4e-18
Identities = 65/159 (40%), Positives = 82/159 (50%), Gaps = 15/159 (9%)
Frame = +1
Query: 937 LLIVYQIFGNGYRSGLSYLLITTPMWFMVGTWLYAPFLFNPSGFEWQKI---VDDWTDWN 993
LL+VY+IFG+ YRS ++Y+LIT MWFMVGTWL+APFLFNPSGFEWQK +D +
Sbjct: 1 LLVVYEIFGHSYRSTVAYILITASMWFMVGTWLFAPFLFNPSGFEWQKNC**LDRLE*MD 180
Query: 994 -----------KWISIRGGIGVPPEKSWESWWEEEQEHLKYSGMRGIIAEILLSLRFFI- 1041
K + I G G ++ +W YSG II L + I
Sbjct: 181 **SRGYRCTA*KKLGILVGRGT---RASPIFWNAG----NYSGDTVIIEIFYLPVWSGIS 339
Query: 1042 YQYGLVYHLNFTKSTKSVLPTGLWHIMVGDLSNISYLED 1080
+Y H F +GLWH MVGD+ NI ED
Sbjct: 340 LKYYEERHKKF---------SGLWHFMVGDIRNIVCDED 429
>CO983301
Length = 825
Score = 373 bits (957), Expect = e-103
Identities = 167/266 (62%), Positives = 212/266 (78%)
Frame = -2
Query: 795 ITVLTVYVFLYGRLYLVLSGLEEGLSTQKAIRDNKPLQVALASQSFVQIGFLMALPMLME 854
+TVL VY FLYGRLY+VLSG+E + I +K L+ A+A+QS VQ+G L+ LPM+ME
Sbjct: 815 VTVLIVYAFLYGRLYMVLSGVEREILQSLNIHQSKALEEAMATQSVVQLGLLLLLPMVME 636
Query: 855 IGLERGFRTALSEFILMQLQLAPVFFTFSLGTKTHYYGRTLLHGGAKYRPTGRGFVVFHA 914
IGLERGFRTA+++FI+MQLQLA VFFTF LGTK HYYGRTLLHGG+KYRPTGRG +VFH
Sbjct: 635 IGLERGFRTAVADFIIMQLQLASVFFTFQLGTKAHYYGRTLLHGGSKYRPTGRGLIVFHV 456
Query: 915 KFADNYRLYSRSHFVKGIELMILLIVYQIFGNGYRSGLSYLLITTPMWFMVGTWLYAPFL 974
KFADNYR+YSRSHFVKG+E+++LLIVY+++G YRS YL I +WF+ +WL+APFL
Sbjct: 455 KFADNYRMYSRSHFVKGLEILLLLIVYELYGESYRSSHLYLFIIISIWFLATSWLFAPFL 276
Query: 975 FNPSGFEWQKIVDDWTDWNKWISIRGGIGVPPEKSWESWWEEEQEHLKYSGMRGIIAEIL 1034
FNPSGF+ K VDDWTDW +W+ GIG+ ++SWESWW+E+ EHLKYS +RG I EI+
Sbjct: 275 FNPSGFDLLKTVDDWTDWKRWMGYPDGIGISSDRSWESWWDEQNEHLKYSNLRGKIIEII 96
Query: 1035 LSLRFFIYQYGLVYHLNFTKSTKSVL 1060
L+ R F+YQYG+VYH++ K +L
Sbjct: 95 LAFRXFMYQYGIVYHMDKXXHNKDLL 18
>TC227922 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like protein
(Fragment), partial (23%)
Length = 1119
Score = 292 bits (747), Expect = 7e-79
Identities = 148/221 (66%), Positives = 177/221 (79%), Gaps = 9/221 (4%)
Frame = +2
Query: 1000 GGIGVPPEKSWESWWEEEQEHLKYSGMRGIIAEILLSLRFFIYQYGLVYHLNFTKSTKSV 1059
GGIGV EK WESWWE+E EHL++SG RGI EI+LSLRFFIYQYGLVYHL+ T T+SV
Sbjct: 2 GGIGVSAEKIWESWWEKEHEHLRHSGKRGIATEIILSLRFFIYQYGLVYHLSITDKTQSV 181
Query: 1060 LPTGL-WHIM---VGDLSNISY-----LEDFQLVFRLMKGLVFVTFVSILVTMIALAHMT 1110
L GL W I+ +G + +S D+QL+FRL+ G +F+TF++I + +IA+A MT
Sbjct: 182 LVYGLSWMIIFVILGLMKGVSVGRRRLSADYQLLFRLIVGSIFLTFLAIFIILIAVAKMT 361
Query: 1111 LQDIVVCILAFMPTGWGMLQIAQALKPLVRRGGFWESVKTLARGYEVIMGLLLFTPVAFL 1170
++DI+VCILA MPTGWG+L IAQA KPL+++ FW SV+ LARGYEVIMGLLLFTPVAFL
Sbjct: 362 IKDIIVCILAVMPTGWGILLIAQACKPLIKKTWFWGSVRALARGYEVIMGLLLFTPVAFL 541
Query: 1171 AWFPFVSEFQTRMLFNQAFSRGLQISRILGGQRKGRSSRNK 1211
AWFPFVSEFQTRMLFNQAFSRGLQISRILGGQ RSS +K
Sbjct: 542 AWFPFVSEFQTRMLFNQAFSRGLQISRILGGQSNERSSNHK 664
>TC218628 similar to UP|Q9LWU7 (Q9LWU7) ESTs AU033035(S1515), partial (13%)
Length = 1115
Score = 277 bits (708), Expect = 2e-74
Identities = 130/284 (45%), Positives = 192/284 (66%), Gaps = 8/284 (2%)
Frame = +1
Query: 928 FVKGIELMILLIVYQIFGNGYRSGLSYLLITTPMWFMVGTWLYAPFLFNPSGFEWQKIVD 987
FVK +E+ +LLIVY +G ++Y+L+T WF+V +WL+AP++FNPSGFEWQK V+
Sbjct: 1 FVKALEVALLLIVYIAYGYAEGGAVTYVLLTLSSWFLVISWLFAPYIFNPSGFEWQKTVE 180
Query: 988 DWTDWNKWISIRGGIGVPPEKSWESWWEEEQEHLKYSGMRGIIAEILLSLRFFIYQYGLV 1047
D+ DW W+ +GG+GV + SWESWW+EEQ H++ +RG I E +LS RFF++QYG+V
Sbjct: 181 DFDDWTSWLLYKGGVGVKGDNSWESWWDEEQMHIQT--LRGRILETILSARFFLFQYGVV 354
Query: 1048 YHLNFTKSTKSVLPTGL-WHIMVGDLSNISYLE-------DFQLVFRLMKGLVFVTFVSI 1099
Y L+ T + S+ G W ++VG + +FQLV R +G+ + V+
Sbjct: 355 YKLHLTGNNTSLAIYGFSWAVLVGIVLIFKIFAYSPKKAANFQLVLRFAQGVASIGLVAA 534
Query: 1100 LVTMIALAHMTLQDIVVCILAFMPTGWGMLQIAQALKPLVRRGGFWESVKTLARGYEVIM 1159
+ ++A +++ D+ ILAF+PTGWG+L +A A K +V G W+SV+ AR Y+ M
Sbjct: 535 VCLVVAFTQLSIADLFASILAFIPTGWGILSLAIAWKKIVWSLGMWDSVREFARMYDAGM 714
Query: 1160 GLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISRILGGQR 1203
G+++F P+AFL+WFPF+S FQ+R+LFNQAFSRGL+IS IL G +
Sbjct: 715 GMIIFAPIAFLSWFPFISTFQSRLLFNQAFSRGLEISIILAGNK 846
>TC222995 weakly similar to UP|GLS2_YEAST (P40989) 1,3-beta-glucan synthase
component GLS2 (1,3-beta-D-glucan-UDP
glucosyltransferase) , partial (3%)
Length = 490
Score = 274 bits (700), Expect = 2e-73
Identities = 129/164 (78%), Positives = 150/164 (90%)
Frame = +1
Query: 654 SLAWFMSNQETSFVTIGQRLLANPLRVRFHYGHPDVFDRIFHLTRGGVSKASKVINLSED 713
SLA FMSNQETSFVT+GQR+LANPL+VR HYGHPDVFDRIFH+TRGG+SKAS+VIN+SED
Sbjct: 1 SLASFMSNQETSFVTLGQRVLANPLKVRMHYGHPDVFDRIFHVTRGGISKASRVINISED 180
Query: 714 IFAGFNSTLREGNVTHHEYIQVGKGRDVGLNQISMFEAKIANGNGEQTLSRDVYRLGHRF 773
I++GFNSTLR+GN+THHEYIQVGKGRDVGLNQI++FE K++ GNGEQ LSRDVYRLG F
Sbjct: 181 IYSGFNSTLRQGNITHHEYIQVGKGRDVGLNQIALFEGKVSGGNGEQVLSRDVYRLGQLF 360
Query: 774 DFFRMLSCYFTTIGFYFSTLQITVLTVYVFLYGRLYLVLSGLEE 817
DFFRMLS YFTT+G+YF T+ +TVLTVY FLYG+ YL LSG+ E
Sbjct: 361 DFFRMLSFYFTTVGYYFCTM-LTVLTVYAFLYGKAYLALSGVGE 489
>BM085974 homologue to PIR|T49914|T4 callose synthase catalytic subunit-like
protein - Arabidopsis thaliana, partial (7%)
Length = 427
Score = 273 bits (699), Expect = 3e-73
Identities = 139/143 (97%), Positives = 140/143 (97%)
Frame = +1
Query: 715 FAGFNSTLREGNVTHHEYIQVGKGRDVGLNQISMFEAKIANGNGEQTLSRDVYRLGHRFD 774
F GFNSTLREGNVTHHEYIQVGKGRDVGLNQISMFEAKIANGNGEQTLSRDVYRLGHRFD
Sbjct: 1 FLGFNSTLREGNVTHHEYIQVGKGRDVGLNQISMFEAKIANGNGEQTLSRDVYRLGHRFD 180
Query: 775 FFRMLSCYFTTIGFYFSTLQITVLTVYVFLYGRLYLVLSGLEEGLSTQKAIRDNKPLQVA 834
FFRMLSCYFTT+GFYFSTL ITVLTVYVFLYGRLYLVLSGLEEGLSTQKAIRDNKPLQVA
Sbjct: 181 FFRMLSCYFTTVGFYFSTL-ITVLTVYVFLYGRLYLVLSGLEEGLSTQKAIRDNKPLQVA 357
Query: 835 LASQSFVQIGFLMALPMLMEIGL 857
LASQSFVQIG LMALPMLMEIGL
Sbjct: 358 LASQSFVQIGVLMALPMLMEIGL 426
>TC217078
Length = 1010
Score = 272 bits (695), Expect = 7e-73
Identities = 130/258 (50%), Positives = 184/258 (70%), Gaps = 8/258 (3%)
Frame = +3
Query: 952 LSYLLITTPMWFMVGTWLYAPFLFNPSGFEWQKIVDDWTDWNKWISIRGGIGVPPEKSWE 1011
LSY+L++ WFM +WL+AP+LFNPSGFEWQK+V+D+ DW W+ RGGIGV E+SWE
Sbjct: 3 LSYILLSISSWFMALSWLFAPYLFNPSGFEWQKVVEDFRDWTNWLLYRGGIGVKGEESWE 182
Query: 1012 SWWEEEQEHLKYSGMRGIIAEILLSLRFFIYQYGLVYHLNFTKSTKSVLPTGLWHIMVGD 1071
+WWEEE H++ G R IAE +LSLRFFI+QYG+VY LN ++ S+ GL +++
Sbjct: 183 AWWEEELAHIRSLGSR--IAETILSLRFFIFQYGIVYKLNVKGTSTSLTVYGLSWVVLAV 356
Query: 1072 LSNISYL--------EDFQLVFRLMKGLVFVTFVSILVTMIALAHMTLQDIVVCILAFMP 1123
L + + +FQL+ R ++G+ + ++ LV + L ++L DI +LAF+P
Sbjct: 357 LIILFKVFTFSQKISVNFQLLLRFIQGVSLLVALAGLVVAVILTKLSLPDIFASMLAFIP 536
Query: 1124 TGWGMLQIAQALKPLVRRGGFWESVKTLARGYEVIMGLLLFTPVAFLAWFPFVSEFQTRM 1183
TGWG+L IA A KP+++R G W+SV+++AR Y+ MG+L+F P+AF +WFPFVS FQTR+
Sbjct: 537 TGWGILSIAAAWKPVMKRLGLWKSVRSIARLYDAGMGMLIFVPIAFFSWFPFVSTFQTRL 716
Query: 1184 LFNQAFSRGLQISRILGG 1201
+FNQAFSRGL+IS IL G
Sbjct: 717 MFNQAFSRGLEISLILAG 770
>BE660208
Length = 720
Score = 246 bits (627), Expect(2) = 3e-71
Identities = 132/199 (66%), Positives = 152/199 (76%)
Frame = +2
Query: 507 AARAQDILRLMARYPSLRVAYIDEVEEPSKERPKRISKVYYSCLVKAMPKSSSPSETEPE 566
A A DI L+ R +LRVA+I V+E + + SKV+YS LVKA
Sbjct: 5 APEAADIALLLQRNEALRVAFI-HVDESTTD--VNTSKVFYSKLVKADINGK-------- 151
Query: 567 QCLDQVIYKIKLPGPAILGEGKPENQNHAIMFTRGEGLQTIDMNQDNYMEEALKMRNLLQ 626
DQ IY IKLPG LGEGKPENQNHAI+FTRGE +QTIDMNQDNY+EEA+KMRNLL+
Sbjct: 152 ---DQEIYSIKLPGDPKLGEGKPENQNHAIIFTRGEAVQTIDMNQDNYLEEAMKMRNLLE 322
Query: 627 EFLKKHDGVRYPSILGLREHIFTGSVSSLAWFMSNQETSFVTIGQRLLANPLRVRFHYGH 686
EF H G+R PSILG+REH+FTGSVSSLAWFMSNQETSFVT+ QR+LANPL+VR HYGH
Sbjct: 323 EFHANH-GLRPPSILGVREHVFTGSVSSLAWFMSNQETSFVTLAQRVLANPLKVRMHYGH 499
Query: 687 PDVFDRIFHLTRGGVSKAS 705
PDVFDRIFH+TRGG+SKAS
Sbjct: 500 PDVFDRIFHITRGGISKAS 556
Score = 43.1 bits (100), Expect(2) = 3e-71
Identities = 26/50 (52%), Positives = 32/50 (64%), Gaps = 2/50 (4%)
Frame = +1
Query: 710 LSEDIFAGFNSTLREGNVTH-HEYIQVGKGRDV-GLNQISMFEAKIANGN 757
L +DI+AGFNSTLR G +H IQVGK G QI++ E K+A GN
Sbjct: 571 LVKDIYAGFNSTLRXGKWSHIMNTIQVGKXXGCWG*TQIALXEGKVAXGN 720
>TC217351 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like protein
(Fragment), partial (13%)
Length = 601
Score = 209 bits (532), Expect = 6e-54
Identities = 106/126 (84%), Positives = 114/126 (90%)
Frame = +3
Query: 1086 RLMKGLVFVTFVSILVTMIALAHMTLQDIVVCILAFMPTGWGMLQIAQALKPLVRRGGFW 1145
RL+KG++ TFVSILV + A+ L DIVVCILAFMPTGWGMLQIAQALKP+VRR GFW
Sbjct: 3 RLIKGMIXXTFVSILVI*LLXAYXXL-DIVVCILAFMPTGWGMLQIAQALKPVVRRAGFW 179
Query: 1146 ESVKTLARGYEVIMGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISRILGGQRKG 1205
SVKTLARGYE++MGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISRILGGQRK
Sbjct: 180 GSVKTLARGYEIVMGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISRILGGQRKE 359
Query: 1206 RSSRNK 1211
RSSRNK
Sbjct: 360 RSSRNK 377
>TC227920 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like protein
(Fragment), partial (13%)
Length = 675
Score = 207 bits (526), Expect = 3e-53
Identities = 99/131 (75%), Positives = 117/131 (88%)
Frame = +1
Query: 1081 FQLVFRLMKGLVFVTFVSILVTMIALAHMTLQDIVVCILAFMPTGWGMLQIAQALKPLVR 1140
+QL+FRL++G +F+TF++I + +I LA+MT++DI+VCILA MPTGWGML IAQA KPL+
Sbjct: 1 YQLLFRLIEGSIFLTFLAIFIILILLANMTIKDIIVCILAVMPTGWGMLLIAQACKPLIE 180
Query: 1141 RGGFWESVKTLARGYEVIMGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISRILG 1200
+ GFW SV+ LARGYEVIMGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISRILG
Sbjct: 181 KTGFWGSVRALARGYEVIMGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISRILG 360
Query: 1201 GQRKGRSSRNK 1211
GQR RSS +K
Sbjct: 361 GQRSERSSNHK 393
>BE058482
Length = 430
Score = 181 bits (459), Expect = 2e-45
Identities = 87/93 (93%), Positives = 91/93 (97%)
Frame = +1
Query: 1112 QDIVVCILAFMPTGWGMLQIAQALKPLVRRGGFWESVKTLARGYEVIMGLLLFTPVAFLA 1171
+DIVVCILAFMPTGWGMLQIAQALKP+VRR GFW SVKTLARGYE++MGLLLFTPVAFLA
Sbjct: 148 RDIVVCILAFMPTGWGMLQIAQALKPVVRRAGFWGSVKTLARGYEIVMGLLLFTPVAFLA 327
Query: 1172 WFPFVSEFQTRMLFNQAFSRGLQISRILGGQRK 1204
WFPFVSEFQTRMLFNQAFSRGLQISRILGGQRK
Sbjct: 328 WFPFVSEFQTRMLFNQAFSRGLQISRILGGQRK 426
>TC208781 similar to UP|Q9SHJ3 (Q9SHJ3) F12K11.17, partial (8%)
Length = 828
Score = 179 bits (454), Expect = 7e-45
Identities = 88/181 (48%), Positives = 124/181 (67%), Gaps = 9/181 (4%)
Frame = +1
Query: 1032 EILLSLRFFIYQYGLVYHLNFTKSTKSVLPTGL-WHIMVGDLSNISYLE--------DFQ 1082
EI+L+ RFF+YQYG VYH++ T K +L GL W V L + + DFQ
Sbjct: 4 EIVLAFRFFMYQYGXVYHMDITHHNKDLLVFGLSWAXXVIILIVLKMVSMGRRRFGTDFQ 183
Query: 1083 LVFRLMKGLVFVTFVSILVTMIALAHMTLQDIVVCILAFMPTGWGMLQIAQALKPLVRRG 1142
L+FR++K L+F+ F+S++ + + +T+ D+ I+AFMP+GW ++ IAQA K ++
Sbjct: 184 LMFRILKALLFLGFLSVMTVLFVVCGLTIADLFAAIIAFMPSGWAIILIAQACKVCLKGA 363
Query: 1143 GFWESVKTLARGYEVIMGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISRILGGQ 1202
W+SVK L+R YE +MGL++F P A L+WFPFVSEFQTR+LFNQAFSRGLQIS IL G+
Sbjct: 364 KLWDSVKELSRAYEYVMGLIIFLPTAILSWFPFVSEFQTRLLFNQAFSRGLQISMILAGK 543
Query: 1203 R 1203
+
Sbjct: 544 K 546
>BE059506 similar to PIR|F86200|F8 protein F12K11.17 [imported] - Arabidopsis
thaliana, partial (5%)
Length = 320
Score = 171 bits (433), Expect = 2e-42
Identities = 71/104 (68%), Positives = 87/104 (83%)
Frame = +1
Query: 912 FHAKFADNYRLYSRSHFVKGIELMILLIVYQIFGNGYRSGLSYLLITTPMWFMVGTWLYA 971
FHAKFADNYR+YSRSHFVKG+E++ILLIVY+++G+ YRS YL IT MWF+ WL+A
Sbjct: 7 FHAKFADNYRMYSRSHFVKGLEILILLIVYEVYGSSYRSSHLYLFITISMWFVATYWLFA 186
Query: 972 PFLFNPSGFEWQKIVDDWTDWNKWISIRGGIGVPPEKSWESWWE 1015
PFLFNPSGF+ QK VDDWTDW +W+ RGGIG+ +KSWESWW+
Sbjct: 187 PFLFNPSGFDGQKTVDDWTDWKRWMGNRGGIGILSDKSWESWWD 318
>BM523215
Length = 433
Score = 169 bits (427), Expect = 9e-42
Identities = 85/140 (60%), Positives = 103/140 (72%)
Frame = +3
Query: 768 RLGHRFDFFRMLSCYFTTIGFYFSTLQITVLTVYVFLYGRLYLVLSGLEEGLSTQKAIRD 827
RLG FDFFRMLS +FTT+G+Y T+ +TVLTVY+FLYGR YL SGL+E +S + ++
Sbjct: 15 RLGQLFDFFRMLSFFFTTVGYYVCTM-MTVLTVYIFLYGRAYLAFSGLDEAVSEKAKLQG 191
Query: 828 NKPLQVALASQSFVQIGFLMALPMLMEIGLERGFRTALSEFILMQLQLAPVFFTFSLGTK 887
N L AL +Q VQIG A+PM+M LE G A+ FI MQLQL VFFTFSLGT+
Sbjct: 192 NTALDAALNAQFLVQIGVFTAVPMIMGFILELGLLKAVFSFITMQLQLCSVFFTFSLGTR 371
Query: 888 THYYGRTLLHGGAKYRPTGR 907
THY+GRT+LHGGAKYR TGR
Sbjct: 372 THYFGRTILHGGAKYRATGR 431
>BM187676
Length = 437
Score = 97.8 bits (242), Expect(2) = 4e-38
Identities = 43/69 (62%), Positives = 58/69 (83%)
Frame = +2
Query: 849 LPMLMEIGLERGFRTALSEFILMQLQLAPVFFTFSLGTKTHYYGRTLLHGGAKYRPTGRG 908
+PM++E LE GF A+ +F+ MQLQL+ VF+TFS+GT++H++GRT+LHGGAKYR TGRG
Sbjct: 2 VPMIVENSLEHGFLQAIWDFLTMQLQLSSVFYTFSMGTRSHFFGRTVLHGGAKYRATGRG 181
Query: 909 FVVFHAKFA 917
FVV H +FA
Sbjct: 182 FVVEHKRFA 208
Score = 80.5 bits (197), Expect(2) = 4e-38
Identities = 34/73 (46%), Positives = 49/73 (66%)
Frame = +3
Query: 920 YRLYSRSHFVKGIELMILLIVYQIFGNGYRSGLSYLLITTPMWFMVGTWLYAPFLFNPSG 979
YRL++RSHFVK IEL ++L++Y Y+ +T WF+V +W+ APF+FNPSG
Sbjct: 216 YRLFARSHFVKAIELGLILVIYASHSPVATDTFVYIALTITSWFLVASWIMAPFVFNPSG 395
Query: 980 FEWQKIVDDWTDW 992
F+W K V D+ D+
Sbjct: 396 FDWLKTVYDFDDF 434
>BG508557 weakly similar to GP|9294379|dbj glucan synthase-like protein
{Arabidopsis thaliana}, partial (4%)
Length = 437
Score = 150 bits (379), Expect = 3e-36
Identities = 62/105 (59%), Positives = 79/105 (75%)
Frame = +1
Query: 940 VYQIFGNGYRSGLSYLLITTPMWFMVGTWLYAPFLFNPSGFEWQKIVDDWTDWNKWISIR 999
VY +F Y+S ++Y+LIT +WFM TWL APFLFNP+GF W K VDDW +WNKWI +
Sbjct: 1 VYNMFRRSYQSSMAYVLITYAIWFMSLTWLCAPFLFNPAGFSWTKTVDDWKEWNKWIRQQ 180
Query: 1000 GGIGVPPEKSWESWWEEEQEHLKYSGMRGIIAEILLSLRFFIYQY 1044
GGIG+ ++SW SWW +EQ HL++SG + E+LLSLRFFIYQY
Sbjct: 181 GGIGIQQDRSWHSWWHDEQAHLRWSGFGSRLTEVLLSLRFFIYQY 315
>AW133183 weakly similar to PIR|T49914|T49 callose synthase catalytic
subunit-like protein - Arabidopsis thaliana, partial (4%)
Length = 445
Score = 137 bits (345), Expect = 3e-32
Identities = 72/137 (52%), Positives = 87/137 (62%)
Frame = +2
Query: 891 YGRTLLHGGAKYRPTGRGFVVFHAKFADNYRLYSRSHFVKGIELMILLIVYQIFGNGYRS 950
YGRTLLHGGA+Y+ TG G VV +A AD+ L H + IE+MILL+VY I+G+ YR
Sbjct: 2 YGRTLLHGGAEYKSTGSGSVVLYAISADDSGLCYTDHSDQRIEVMILLVVYHIYGHDYR* 181
Query: 951 GLSYLLITTPMWFMVGTWLYAPFLFNPSGFEWQKIVDDWTDWNKWISIRGGIGVPPEKSW 1010
L+Y+ IT MWFMVGTWL AP LFN S EWQ I D T W+S+R G V K +
Sbjct: 182 VLAYI*ITVTMWFMVGTWLVAPLLFNTSRLEWQLIAHD*TYSLTWMSLRVGTCVSSAKRY 361
Query: 1011 ESWWEEEQEHLKYSGMR 1027
ESWW E + SG R
Sbjct: 362 ESWWANGLEPSRCSGKR 412
>BU081888
Length = 425
Score = 129 bits (324), Expect = 8e-30
Identities = 65/140 (46%), Positives = 93/140 (66%), Gaps = 9/140 (6%)
Frame = +3
Query: 1015 EEEQEHLKYSGMRGIIAEILLSLRFFIYQYGLVYHLNFTKSTKSVLPTGL-WHIMVG--- 1070
+EEQEHL+Y+G+ G I E++L+LRFF+YQYG+VYHL+ + KS+ GL W ++V
Sbjct: 3 DEEQEHLQYTGIWGRIWEVILALRFFVYQYGIVYHLHVARGDKSIGVYGLSWLVVVAVIV 182
Query: 1071 -----DLSNISYLEDFQLVFRLMKGLVFVTFVSILVTMIALAHMTLQDIVVCILAFMPTG 1125
+ + ++ DFQL+FRL+K +F+ + IL+ M L T+ DI +LAFMPTG
Sbjct: 183 ILKIVSMGSKTFSADFQLMFRLLKMFLFIGTIVILILMFVLLSFTVGDIFASLLAFMPTG 362
Query: 1126 WGMLQIAQALKPLVRRGGFW 1145
W +QIAQA KPLV+ G W
Sbjct: 363 WAFIQIAQACKPLVKGIGMW 422
>TC220274
Length = 941
Score = 116 bits (290), Expect = 7e-26
Identities = 59/170 (34%), Positives = 97/170 (56%), Gaps = 9/170 (5%)
Frame = +1
Query: 1043 QYGLVYHLNFTKSTKSVLPTGLWHIMVGDLSNI---------SYLEDFQLVFRLMKGLVF 1093
QYG+VY L + S+ L I V +S I Y + +RL++ LV
Sbjct: 4 QYGIVYQLGISDHNTSIAVYLLSWIYVFVVSGIYAVVVYARNKYAAKEHIYYRLVQFLVI 183
Query: 1094 VTFVSILVTMIALAHMTLQDIVVCILAFMPTGWGMLQIAQALKPLVRRGGFWESVKTLAR 1153
+ + ++V ++ DI +LAF+PTGWG++ IAQ +P ++ W+ V ++AR
Sbjct: 184 ILAILVIVGLLEFTKFKFMDIFTSLLAFIPTGWGLISIAQVFRPFLQSTIIWDGVVSVAR 363
Query: 1154 GYEVIMGLLLFTPVAFLAWFPFVSEFQTRMLFNQAFSRGLQISRILGGQR 1203
Y+++ G+++ PVA L+W P QTR+LFN+AFSRGL+I +I+ G++
Sbjct: 364 IYDIMFGVIIMAPVALLSWLPGFQNMQTRILFNEAFSRGLRIFQIVTGKK 513
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 51,482,889
Number of Sequences: 63676
Number of extensions: 731587
Number of successful extensions: 3539
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 3495
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3521
length of query: 1211
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1103
effective length of database: 5,762,624
effective search space: 6356174272
effective search space used: 6356174272
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 64 (29.3 bits)
Medicago: description of AC137603.6


