

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137603.10 + phase: 0
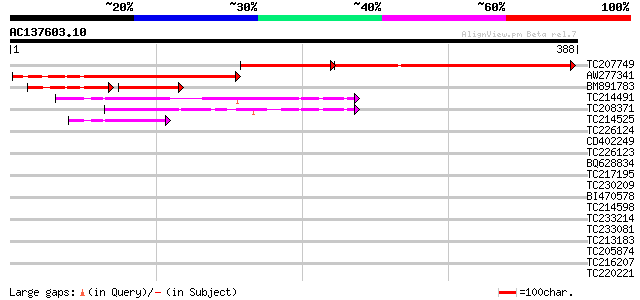
(388 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207749 similar to GB|AAO63411.1|28950975|BT005347 At1g51115 {A... 255 4e-99
AW277341 208 4e-54
BM891783 79 5e-25
TC214491 65 7e-11
TC208371 similar to UP|LIPC_SOLTU (P80471) Light-induced protein... 59 3e-09
TC214525 weakly similar to UP|Q9LU84 (Q9LU84) Arabidopsis thalia... 42 5e-04
TC226124 similar to UP|Q9ZWQ8 (Q9ZWQ8) Homolog to plastid-lipid-... 39 0.005
CD402249 homologue to PIR|S75332|S753 fibrillin - Synechocystis ... 38 0.007
TC226123 similar to UP|O24141 (O24141) Plastid-lipid-Associated ... 37 0.015
BQ628834 35 0.044
TC217195 similar to UP|Q9ZP40 (Q9ZP40) Plastoglobule associated ... 35 0.044
TC230209 34 0.13
BI470578 similar to GP|6448579|emb| nodule inception protein {Lo... 32 0.49
TC214598 similar to UP|P73219 (P73219) Fibrillin, partial (11%) 30 1.4
TC233214 similar to UP|O01940 (O01940) Sno, partial (9%) 29 4.1
TC233081 28 7.0
TC213183 similar to UP|Q6YUW7 (Q6YUW7) Chromosome structural mai... 28 7.0
TC205874 similar to UP|Q945S0 (Q945S0) U2 auxiliary factor small... 28 7.0
TC216207 similar to GB|AAN72183.1|25084133|BT002172 protein kina... 28 7.0
TC220221 28 9.2
>TC207749 similar to GB|AAO63411.1|28950975|BT005347 At1g51115 {Arabidopsis
thaliana;} , partial (44%)
Length = 1069
Score = 255 bits (652), Expect(2) = 4e-99
Identities = 124/165 (75%), Positives = 147/165 (88%)
Frame = +3
Query: 223 KGTTFVLQKQTEPRQKLLTAISSGVGVREAIDKLISLNKNSGEEDPELEEGEWQMIWNSQ 282
+GTTFVLQKQTEPRQ+LLTAISSG G++EAID+LISL +N G+E PELE+GEWQM+WNSQ
Sbjct: 360 QGTTFVLQKQTEPRQRLLTAISSGKGIKEAIDELISLKQNIGQE-PELEDGEWQMMWNSQ 536
Query: 283 TVTDSWLENAANGLMGKQIVEKNGRIKYVVNILLGLKFSMSGIFVKSSPKVYEVTMDDAA 342
TVTDSW+ENA NGLMGKQI+ KNGRIK++V+ILLGLKFSMSG FVK+ VY VTMDDAA
Sbjct: 537 TVTDSWIENAVNGLMGKQIIRKNGRIKFLVDILLGLKFSMSGNFVKTGSGVYGVTMDDAA 716
Query: 343 IIGGPFGYPLEFGKKFILEILFNDGKVRISRGDNEIIFVHARTNA 387
IIGGPFGYPLE FILE+L++D K+R+SRG+N I+FVH RT+A
Sbjct: 717 IIGGPFGYPLELKNNFILELLYSDEKLRVSRGNNSILFVHVRTDA 851
Score = 124 bits (311), Expect(2) = 4e-99
Identities = 59/65 (90%), Positives = 62/65 (94%)
Frame = +1
Query: 159 EAAASIGDGKRILFRFDRAAFSFKFLPFKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRI 218
+AAASI DGKRILFRFDRAAFSFKFLPFKVPYPVPF+LLGDEAKGWLDTTYLS SGNL I
Sbjct: 163 QAAASIEDGKRILFRFDRAAFSFKFLPFKVPYPVPFRLLGDEAKGWLDTTYLSSSGNLHI 342
Query: 219 SRGNK 223
S+GNK
Sbjct: 343 SKGNK 357
>AW277341
Length = 444
Score = 208 bits (529), Expect = 4e-54
Identities = 115/156 (73%), Positives = 126/156 (80%)
Frame = +3
Query: 3 LTLRVVNVNSIGFESCSRCSTFISPNPSNSRFVSAGCSKVEQISIVTEESENSLIQALVG 62
+ L++VN +GF S CS P P RFV SKVEQIS ESENSLI+AL+G
Sbjct: 6 MALKLVN---LGFHSSIPCS----PRPLKDRFVLRS-SKVEQISFT--ESENSLIEALLG 155
Query: 63 IQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSSLIEGRWQLIFTTRPGTASPIQRTF 122
IQGRGRSS QQLNA+ERA+QVLE +GGV DPT S+LIEGRWQLIFTTRPGTASPIQRTF
Sbjct: 156 IQGRGRSSFRQQLNAVERAVQVLERLGGVPDPTKSNLIEGRWQLIFTTRPGTASPIQRTF 335
Query: 123 VGVDFFSVFQEVYLQTNDPRVTNIVSFSDAIGELKV 158
VGVDFFSVFQEVYL+TNDPRV NIVSFSDAIGELKV
Sbjct: 336 VGVDFFSVFQEVYLRTNDPRVCNIVSFSDAIGELKV 443
>BM891783
Length = 424
Score = 79.3 bits (194), Expect(2) = 5e-25
Identities = 37/45 (82%), Positives = 41/45 (90%)
Frame = +2
Query: 75 LNAIERAIQVLEHIGGVSDPTNSSLIEGRWQLIFTTRPGTASPIQ 119
L A+ERA+QVLE +GGV DPT S+LIEGRWQLIFTTRPGTASPIQ
Sbjct: 152 LLAVERAVQVLERLGGVPDPTKSNLIEGRWQLIFTTRPGTASPIQ 286
Score = 53.1 bits (126), Expect(2) = 5e-25
Identities = 35/59 (59%), Positives = 38/59 (64%)
Frame = +1
Query: 13 IGFESCSRCSTFISPNPSNSRFVSAGCSKVEQISIVTEESENSLIQALVGIQGRGRSSS 71
+GF S CS P RFV SKVEQIS ESENSLI+AL+GIQGRGRSSS
Sbjct: 1 LGFHSSIPCSA----RPLKDRFVLRS-SKVEQISFT--ESENSLIEALLGIQGRGRSSS 156
>TC214491
Length = 1149
Score = 64.7 bits (156), Expect = 7e-11
Identities = 57/213 (26%), Positives = 96/213 (44%), Gaps = 5/213 (2%)
Frame = +1
Query: 32 SRFVSAGCSKVEQISIVTEESENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGV 91
S F S K + I+ EE L++A+ + RG ++PQ +++ + LE + +
Sbjct: 199 SFFTSFLKQKGKDAKIIKEE----LLEAIAPLD-RGADATPQDQQTVDQIARELEAVTPI 363
Query: 92 SDPTNSSLIEGRWQLIFTTRPGTASPIQRTFVGVDFFSVFQEVYLQTNDPRVTNIVSFSD 151
+P S+L++G+W+LI+TT + LQT P++ V+
Sbjct: 364 KEPLKSNLLDGKWELIYTT---------------------SQSILQTKRPKLLRSVANYQ 480
Query: 152 AIG-----ELKVEAAASIGDGKRILFRFDRAAFSFKFLPFKVPYPVPFKLLGDEAKGWLD 206
AI +E+ L + + KF FK+ +P K G A+G L+
Sbjct: 481 AINVDTLRAQNMESWPFFNQVTADLTPLNPRKVAVKFDTFKIGGIIPIKAPG-RARGELE 657
Query: 207 TTYLSHSGNLRISRGNKGTTFVLQKQTEPRQKL 239
TYL LR+SRG+KG F+L K +P ++
Sbjct: 658 ITYLDE--ELRVSRGDKGNLFIL-KMVDPSYRV 747
>TC208371 similar to UP|LIPC_SOLTU (P80471) Light-induced protein,
chloroplast precursor (Chloroplastic drought-induced
stress protein CDSP-34), partial (10%)
Length = 961
Score = 59.3 bits (142), Expect = 3e-09
Identities = 49/176 (27%), Positives = 85/176 (47%), Gaps = 2/176 (1%)
Frame = +2
Query: 66 RGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSSLIEGRWQLIFTTRPGTASPIQRTFVGV 125
RG ++P+ +++ + LE + V +P S L+ G+W+L +TT QR +
Sbjct: 263 RGAEATPEDQQRVDQIARKLEAVNPVKEPLKSGLLNGKWELFYTTSQSILQ-TQRPKLLR 439
Query: 126 DFFSVFQEVYLQTNDPRVTNIVSFSDAIGELKVEAAASIG--DGKRILFRFDRAAFSFKF 183
++Q + + T R NI ++ +A A++ + KR+ +FD
Sbjct: 440 PNGKIYQAINVDT--LRAQNIETW-----PFYNQATANLVPLNSKRVAVKFDF------- 577
Query: 184 LPFKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQKL 239
FK+ +P K G +G L+ TYL +LRISRGN+G F+L K +P ++
Sbjct: 578 --FKIASLIPIKSPG-SGRGQLEITYLDE--DLRISRGNRGNLFIL-KMVDPSYRV 727
>TC214525 weakly similar to UP|Q9LU84 (Q9LU84) Arabidopsis thaliana genomic
DNA, chromosome 3, P1 clone: MPE11, partial (20%)
Length = 458
Score = 42.0 bits (97), Expect = 5e-04
Identities = 20/70 (28%), Positives = 41/70 (58%)
Frame = +1
Query: 41 KVEQISIVTEESENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSSLI 100
K I+ EE +++A+ + RG ++PQ I++ + LE + + +P ++L+
Sbjct: 205 KAIDAKIIKEE----MLEAIAPLD-RGADATPQDQQTIDQIARELEAVTPIKEPLKTNLL 369
Query: 101 EGRWQLIFTT 110
+G+W+LI+TT
Sbjct: 370 DGKWELIYTT 399
>TC226124 similar to UP|Q9ZWQ8 (Q9ZWQ8) Homolog to plastid-lipid-associated
protein, partial (74%)
Length = 1242
Score = 38.5 bits (88), Expect = 0.005
Identities = 52/226 (23%), Positives = 87/226 (38%), Gaps = 42/226 (18%)
Frame = +3
Query: 49 TEESENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNS-SLIEGRWQLI 107
TE+ + +L+ + G +++S + +E Q LE PT++ +L+ G+W L
Sbjct: 372 TEKLKKALVDSFYGTDLGLKATSETRAEIVELITQ-LEAKNPNPAPTDALTLLNGKWILA 548
Query: 108 FTTRPGTASPIQR-TFVGVDFFSVFQEVYLQTNDPRVTNIVSFSDAIGELKVEAAAS--I 164
+T+ G + T V + Q + T + V N V F+ + + A +
Sbjct: 549 YTSFAGLFPLLSSGTLPLVKVEEISQTI--DTLNFTVQNSVQFAGPLATTSISTNAKFDV 722
Query: 165 GDGKRILFRFDRAAFSFKFLPFKVPYPVPFKLLGDE------------------------ 200
KR+ +F+ L + P +LLG +
Sbjct: 723 RSPKRVQIKFEEGIIGTPQLTDSLEIPENVELLGQKIDLTPFKGILTSVQDTASSVVKTI 902
Query: 201 --------------AKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQ 232
A+ WL TTYL LRISRG+ G+ FVL K+
Sbjct: 903 SSRPPLKIPISNSNAQSWLLTTYLDEE--LRISRGDGGSVFVLIKE 1034
>CD402249 homologue to PIR|S75332|S753 fibrillin - Synechocystis sp. (strain
PCC 6803), partial (11%)
Length = 360
Score = 38.1 bits (87), Expect = 0.007
Identities = 24/58 (41%), Positives = 34/58 (58%)
Frame = -1
Query: 182 KFLPFKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQKL 239
KF FK+ +P K G +G L+ TYL +LRISRGN+G F+L K +P ++
Sbjct: 324 KFDFFKIASLIPIKSPGS-GRGQLEITYLDE--DLRISRGNRGNLFIL-KMVDPSYRV 163
>TC226123 similar to UP|O24141 (O24141) Plastid-lipid-Associated Protein
(Fragment), partial (68%)
Length = 833
Score = 37.0 bits (84), Expect = 0.015
Identities = 23/58 (39%), Positives = 30/58 (51%)
Frame = +1
Query: 175 DRAAFSFKFLPFKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQ 232
D A K + + P +P A+ WL TTYL LRISRG+ G+ FVL K+
Sbjct: 376 DTATSVVKTISSRPPLKIPIS--NSNAQSWLLTTYLDEE--LRISRGDGGSVFVLIKE 537
>BQ628834
Length = 304
Score = 35.4 bits (80), Expect = 0.044
Identities = 14/24 (58%), Positives = 20/24 (83%)
Frame = +1
Query: 364 FNDGKVRISRGDNEIIFVHARTNA 387
++D K+R+SRG N +IFVH RT+A
Sbjct: 67 YSDEKLRVSRGINNVIFVHVRTDA 138
>TC217195 similar to UP|Q9ZP40 (Q9ZP40) Plastoglobule associated protein PG1
precursor, partial (96%)
Length = 1388
Score = 35.4 bits (80), Expect = 0.044
Identities = 22/51 (43%), Positives = 28/51 (54%)
Frame = +3
Query: 185 PFKVPYPVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEP 235
P K+P P G+ WL TTYL +LRISRG+ G FVL ++ P
Sbjct: 972 PLKIPIP------GERTSSWLLTTYLDK--DLRISRGD-GGLFVLAREGSP 1097
>TC230209
Length = 899
Score = 33.9 bits (76), Expect = 0.13
Identities = 24/87 (27%), Positives = 41/87 (46%), Gaps = 5/87 (5%)
Frame = +3
Query: 91 VSDPTNSSLIEGRWQLIFTTRPGTASPIQRTFVGVDFFSVFQEVYLQTNDPRVTN----- 145
VS+P LI G W + + +RP + R+ +G FF+ Q V + V N
Sbjct: 303 VSEPVKCPLIFGEWDVAYCSRPTSPGGGYRSAIGRLFFNTKQMVQVVEAPDIVRNKVSLS 482
Query: 146 IVSFSDAIGELKVEAAASIGDGKRILF 172
++SF D L+ + A G+ +++F
Sbjct: 483 VLSFLDVEVSLQGKLKALDGEWIQVIF 563
>BI470578 similar to GP|6448579|emb| nodule inception protein {Lotus
japonicus}, partial (3%)
Length = 381
Score = 32.0 bits (71), Expect = 0.49
Identities = 15/55 (27%), Positives = 28/55 (50%)
Frame = -2
Query: 65 GRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNSSLIEGRWQLIFTTRPGTASPIQ 119
G SS P + ++E + Q++ +SDP S ++ RW + PG +S ++
Sbjct: 176 GEANSSKPNTMASMETSAQIIFQEESLSDPEISLMVGKRWWIGPRENPGPSSSVK 12
>TC214598 similar to UP|P73219 (P73219) Fibrillin, partial (11%)
Length = 524
Score = 30.4 bits (67), Expect = 1.4
Identities = 17/39 (43%), Positives = 24/39 (60%)
Frame = +1
Query: 201 AKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQKL 239
A+G L+ TYL LR SRG+KG F+L K +P ++
Sbjct: 16 ARGELEITYLDEE--LRXSRGDKGNLFIL-KMVDPSYRV 123
>TC233214 similar to UP|O01940 (O01940) Sno, partial (9%)
Length = 702
Score = 28.9 bits (63), Expect = 4.1
Identities = 14/30 (46%), Positives = 15/30 (49%)
Frame = +2
Query: 11 NSIGFESCSRCSTFISPNPSNSRFVSAGCS 40
N GF +C FIS NSRF AG S
Sbjct: 602 NGAGFSACGTSGMFISQTRYNSRFYCAGKS 691
>TC233081
Length = 642
Score = 28.1 bits (61), Expect = 7.0
Identities = 23/79 (29%), Positives = 34/79 (42%), Gaps = 2/79 (2%)
Frame = +1
Query: 258 SLNKNSGEEDPELEEGEW--QMIWNSQTVTDSWLENAANGLMGKQIVEKNGRIKYVVNIL 315
SL +N GE+D E GEW +++ N Q V N L+G NG
Sbjct: 322 SLAQNYGEDDKESGSGEWVDKVMVNKQDV------NKTENLLGCWQAANNG--------- 456
Query: 316 LGLKFSMSGIFVKSSPKVY 334
L + +++ SPK+Y
Sbjct: 457 -NLSEAFYQKYIEDSPKMY 510
>TC213183 similar to UP|Q6YUW7 (Q6YUW7) Chromosome structural maintenance
protein-like, partial (3%)
Length = 517
Score = 28.1 bits (61), Expect = 7.0
Identities = 27/89 (30%), Positives = 42/89 (46%), Gaps = 8/89 (8%)
Frame = -3
Query: 10 VNSIGFESCSRCSTFISPNPSNSRFVSAGCSKVEQ-ISI-VTEESENSLIQALVGIQGRG 67
V+++ S+ + P PS+S +G + E+ IS+ VT + G++G
Sbjct: 476 VSNLATSFAKVASSSLKPLPSSSPLPPSGTTVEERAISLSVTSPKRPWISSEEGGVRGET 297
Query: 68 RSSSPQQLNAIERAIQV------LEHIGG 90
S P QL +ERAIQ+ L H GG
Sbjct: 296 SSEKPSQL--VERAIQLGTLSLELGHEGG 216
>TC205874 similar to UP|Q945S0 (Q945S0) U2 auxiliary factor small subunit,
partial (8%)
Length = 667
Score = 28.1 bits (61), Expect = 7.0
Identities = 24/82 (29%), Positives = 39/82 (47%), Gaps = 5/82 (6%)
Frame = +2
Query: 203 GWLDTTYLSHSGNLRISRG----NKGTTFVLQKQTEPRQKLLTAISSGVGVREAIDK-LI 257
G L + + G L++S G NKG T QK + I + V + A++ +I
Sbjct: 77 GVLRSEKIVQKGELKLSNGTGRRNKGKLV-----TRTIQKAMMIIKNRVLHKMAVNLVII 241
Query: 258 SLNKNSGEEDPELEEGEWQMIW 279
+ N+ E P++ EWQ+IW
Sbjct: 242 RYSNNNVEVWPKICYQEWQVIW 307
>TC216207 similar to GB|AAN72183.1|25084133|BT002172 protein kinase-like
protein {Arabidopsis thaliana;} , partial (77%)
Length = 1503
Score = 28.1 bits (61), Expect = 7.0
Identities = 15/39 (38%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Frame = -1
Query: 279 WNSQTVTDSWLEN--AANGLMGKQIVEKNGRIKYVVNIL 315
WN TVT SW + ++ + +V +NG IKYV+ I+
Sbjct: 1047 WNCTTVTCSWNWR*WREHYILLRLLVLQNGPIKYVIKII 931
>TC220221
Length = 638
Score = 27.7 bits (60), Expect = 9.2
Identities = 21/59 (35%), Positives = 30/59 (50%), Gaps = 3/59 (5%)
Frame = +3
Query: 287 SWLENAANGLMGKQ---IVEKNGRIKYVVNILLGLKFSMSGIFVKSSPKVYEVTMDDAA 342
SWL + L K IVE NGRI ++LLG ++ + G S +E+ +D AA
Sbjct: 387 SWLVQLLSSLNLKTDNGIVESNGRIARAGSMLLG-QYWLRGFISLLSRSDWEIVIDAAA 560
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,070,293
Number of Sequences: 63676
Number of extensions: 187147
Number of successful extensions: 959
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 940
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 953
length of query: 388
length of database: 12,639,632
effective HSP length: 99
effective length of query: 289
effective length of database: 6,335,708
effective search space: 1831019612
effective search space used: 1831019612
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137603.10


