

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137602.5 - phase: 0
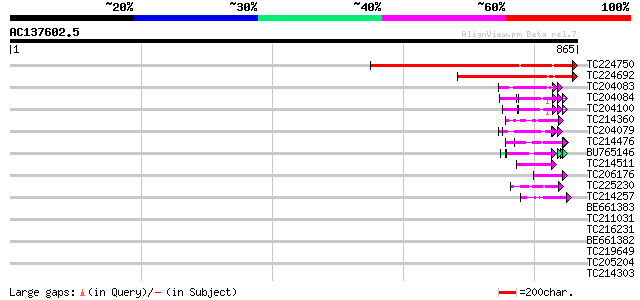
(865 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC224750 similar to UP|IF38_MEDTR (Q9XHM1) Eukaryotic translatio... 496 e-140
TC224692 similar to UP|IF38_MEDTR (Q9XHM1) Eukaryotic translatio... 273 3e-73
TC204083 similar to UP|O48567 (O48567) Glycine-rich RNA-binding ... 55 1e-07
TC204084 UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein, co... 51 3e-06
TC204100 homologue to UP|Q93WA5 (Q93WA5) Ribosomal protein L32, ... 50 4e-06
TC214360 similar to UP|Q9MBF3 (Q9MBF3) Glycine-rich RNA-binding ... 49 7e-06
TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding ... 49 1e-05
TC214476 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 48 2e-05
BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvo... 47 4e-05
TC214511 weakly similar to PIR|C85356|C85356 glycine-rich protei... 45 1e-04
TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein... 44 3e-04
TC225230 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein ... 44 3e-04
TC214257 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 44 4e-04
BE661383 similar to GP|8778710|gb| T1N15.2 {Arabidopsis thaliana... 42 0.001
TC211031 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 42 0.001
TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (G... 42 0.002
BE661382 similar to GP|8778710|gb| T1N15.2 {Arabidopsis thaliana... 42 0.002
TC219649 weakly similar to UP|Q95JD1 (Q95JD1) Basic proline-rich... 41 0.002
TC205204 homologue to UP|Q43558 (Q43558) Proline rich protein pr... 41 0.003
TC214303 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, pa... 41 0.003
>TC224750 similar to UP|IF38_MEDTR (Q9XHM1) Eukaryotic translation initiation
factor 3 subunit 8 (eIF3 p110) (eIF3c), partial (31%)
Length = 1263
Score = 496 bits (1276), Expect = e-140
Identities = 259/318 (81%), Positives = 277/318 (86%), Gaps = 3/318 (0%)
Frame = +2
Query: 551 RQMPYHMHINLELLESVHLTSAMLLEVPNMAANVHDAKRKIISKNFRRLLEVSEKQTFTG 610
RQMPYHMHINLELLE+VHL SAMLLEVPNMAANVHDAKRK+ISK FRRLLEVS++QTFTG
Sbjct: 2 RQMPYHMHINLELLETVHLVSAMLLEVPNMAANVHDAKRKLISKTFRRLLEVSDRQTFTG 181
Query: 611 PPETVRDHVMAATRVLINGDFQKAFDIIASLEVWKFVKNRDTVLEMLKDKIKEEALRTYL 670
PPE VRDHVMAATR L GDFQKAFDII SL+VWKFV+NRDTVLEMLKDKIKEEALRTYL
Sbjct: 182 PPENVRDHVMAATRFLSKGDFQKAFDIIVSLDVWKFVRNRDTVLEMLKDKIKEEALRTYL 361
Query: 671 FTFSSSYDSLSVDQLTNFFDLSLPRAHSIVSRMMINEELHASWDQPTGCIIFRNVELSRV 730
FTFSSSY+SLS+DQLT FFDL + HSIVSRMMINEELHASWDQPTGCI+F++VE SR+
Sbjct: 362 FTFSSSYESLSLDQLTKFFDLPVSCTHSIVSRMMINEELHASWDQPTGCILFQDVEHSRL 541
Query: 731 QALAFQLTEKLSILAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRW 790
QALAFQLTEKLSILAESNERA+EAR+GGGGLDLP RRRDGQDYAAAAA G G SSGGRW
Sbjct: 542 QALAFQLTEKLSILAESNERATEARIGGGGLDLPLRRRDGQDYAAAAA-GSGTASSGGRW 718
Query: 791 QDLSYSQTRQGSGRAGY---GGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALR 847
QDLS SQ RQ SGRAGY GGR ++ Q G GRG G GYQ S R QGGSALR
Sbjct: 719 QDLSLSQPRQSSGRAGYVGGGGRPMALGQGSGYSRDRSGRG-SGAGYQ-SGRYQGGSALR 892
Query: 848 GPHGDVSTRMVSLRGVRA 865
GPHGDVSTRMVSL+GVRA
Sbjct: 893 GPHGDVSTRMVSLKGVRA 946
>TC224692 similar to UP|IF38_MEDTR (Q9XHM1) Eukaryotic translation initiation
factor 3 subunit 8 (eIF3 p110) (eIF3c), partial (16%)
Length = 709
Score = 273 bits (697), Expect = 3e-73
Identities = 143/183 (78%), Positives = 156/183 (85%), Gaps = 1/183 (0%)
Frame = +1
Query: 684 QLTNFFDLSLPRAHSIVSRMMINEELHASWDQPTGCIIFRNVELSRVQALAFQLTEKLSI 743
+LT FFDLS+ R HSIVSRMMINEELHASWDQPTGCI+F++VE SR+QALAFQLTEKLS+
Sbjct: 1 ELTKFFDLSVCRTHSIVSRMMINEELHASWDQPTGCILFQDVEHSRLQALAFQLTEKLSV 180
Query: 744 LAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSG 803
LAESNE+A+EAR+GGGGLDLP RRRDGQDYAAAAA G G SSGGRWQDLS SQ RQGSG
Sbjct: 181 LAESNEKAAEARVGGGGLDLPLRRRDGQDYAAAAAAGSGTASSGGRWQDLSLSQPRQGSG 360
Query: 804 RAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRT-QGGSALRGPHGDVSTRMVSLRG 862
RAGYGGR ++ QA GS GYSRGRG G Y S RT Q GSALRGP GD STRMVSL+G
Sbjct: 361 RAGYGGRPMALGQAAGS-GYSRGRGR--GSYGGSGRTAQRGSALRGPQGDGSTRMVSLKG 531
Query: 863 VRA 865
VRA
Sbjct: 532 VRA 540
>TC204083 similar to UP|O48567 (O48567) Glycine-rich RNA-binding protein,
partial (95%)
Length = 1019
Score = 55.1 bits (131), Expect = 1e-07
Identities = 39/103 (37%), Positives = 47/103 (44%), Gaps = 6/103 (5%)
Frame = +2
Query: 747 SNERASEARLGGGGLDLPPRR---RDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSG 803
S + +A G G DL R + Q A+ GGGGG SGG + S G G
Sbjct: 209 SEQSMKDAIAGMNGQDLDGRNITVNEAQTRASRGGGGGGGFGSGGGYNRGSGGYGGGGGG 388
Query: 804 RAGYGGR---ALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
GYGGR + N GG GGY GR G GG S ++GG
Sbjct: 389 -GGYGGRREGGYNRNGGGGGGGYGGGRDRGYGGDGGSRYSRGG 514
Score = 43.1 bits (100), Expect = 5e-04
Identities = 33/91 (36%), Positives = 41/91 (44%)
Frame = +2
Query: 746 ESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRA 805
E+ RAS GGGG G + + GGGGG G ++ Y++ G G
Sbjct: 284 EAQTRASRGGGGGGGFG----SGGGYNRGSGGYGGGGGGGGYGGRREGGYNRNGGGGGGG 451
Query: 806 GYGGRALSFNQAGGSGGYSRGRGMGGGGYQN 836
GGR + GGS YSRG G GGG N
Sbjct: 452 YGGGRDRGYGGDGGS-RYSRG-GEGGGSDGN 538
Score = 37.7 bits (86), Expect = 0.022
Identities = 28/78 (35%), Positives = 33/78 (41%), Gaps = 3/78 (3%)
Frame = +2
Query: 777 AAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRG---MGGGG 833
A G G GR ++ +QTR G G GG G GGY+RG G GGGG
Sbjct: 230 AIAGMNGQDLDGRNITVNEAQTRASRGGGGGGG-------FGSGGGYNRGSGGYGGGGGG 388
Query: 834 YQNSSRTQGGSALRGPHG 851
R +GG G G
Sbjct: 389 GGYGGRREGGYNRNGGGG 442
>TC204084 UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein, complete
Length = 1937
Score = 50.8 bits (120), Expect = 3e-06
Identities = 35/89 (39%), Positives = 39/89 (43%)
Frame = +2
Query: 748 NERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGY 807
NE S GGGG R G Y + GGGGG G R +D Y G G GY
Sbjct: 1463 NEAQSRGGGGGGGGGGGGYNRGGGGYGGRSGGGGGG--GGYRSRDGGYGGGYGGGGGGGY 1636
Query: 808 GGRALSFNQAGGSGGYSRGRGMGGGGYQN 836
GG G GYSRG G GG++N
Sbjct: 1637 GG--------GRDRGYSRGGDGGDGGWRN 1699
Score = 43.5 bits (101), Expect = 4e-04
Identities = 31/82 (37%), Positives = 35/82 (41%), Gaps = 4/82 (4%)
Frame = +2
Query: 774 AAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGG----SGGYSRGRGM 829
A + GGGGG GG + G GYGGR+ GG GGY G G
Sbjct: 1469 AQSRGGGGGGGGGGGGYN----------RGGGGYGGRSGGGGGGGGYRSRDGGYGGGYGG 1618
Query: 830 GGGGYQNSSRTQGGSALRGPHG 851
GGGG R +G S RG G
Sbjct: 1619 GGGGGYGGGRDRGYS--RGGDG 1678
Score = 42.4 bits (98), Expect = 9e-04
Identities = 25/67 (37%), Positives = 34/67 (50%)
Frame = +2
Query: 777 AAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQN 836
A G G + GR ++ +Q+R G G G GG +N+ GG G G G GGGGY++
Sbjct: 1412 AIGAMNGQNLDGRNITVNEAQSRGGGGGGGGGGGG--YNRGGGGYGGRSGGGGGGGGYRS 1585
Query: 837 SSRTQGG 843
GG
Sbjct: 1586 RDGGYGG 1606
Score = 39.7 bits (91), Expect = 0.006
Identities = 29/98 (29%), Positives = 44/98 (44%), Gaps = 1/98 (1%)
Frame = +2
Query: 747 SNERASEARLGG-GGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRA 805
++E++ + +G G +L R + + GGGGG GG + R G G
Sbjct: 1388 ASEQSMKDAIGAMNGQNLDGRNITVNEAQSRGGGGGGGGGGGGYNRGGGGYGGRSGGGGG 1567
Query: 806 GYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
G G R+ G GGY G G G GG ++ ++GG
Sbjct: 1568 GGGYRS---RDGGYGGGYGGGGGGGYGGGRDRGYSRGG 1672
>TC204100 homologue to UP|Q93WA5 (Q93WA5) Ribosomal protein L32, complete
Length = 1145
Score = 50.1 bits (118), Expect = 4e-06
Identities = 33/85 (38%), Positives = 40/85 (46%)
Frame = -3
Query: 752 SEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRA 811
+++R GGGG R G Y + GGGGG G R +D Y G G GYGG
Sbjct: 402 AQSRGGGGGGGGGGYNRGGGGYGGRSGGGGGG--GGYRSRDGGYGGGYGGGGGGGYGG-- 235
Query: 812 LSFNQAGGSGGYSRGRGMGGGGYQN 836
G GYSRG G GG++N
Sbjct: 234 ------GRDRGYSRGGDGGDGGWRN 178
Score = 42.7 bits (99), Expect = 7e-04
Identities = 31/81 (38%), Positives = 34/81 (41%), Gaps = 4/81 (4%)
Frame = -3
Query: 775 AAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGG----SGGYSRGRGMG 830
A + GGGGG GG G GYGGR+ GG GGY G G G
Sbjct: 402 AQSRGGGGGGGGGG-----------YNRGGGGYGGRSGGGGGGGGYRSRDGGYGGGYGGG 256
Query: 831 GGGYQNSSRTQGGSALRGPHG 851
GGG R +G S RG G
Sbjct: 255 GGGGYGGGRDRGYS--RGGDG 199
Score = 42.4 bits (98), Expect = 9e-04
Identities = 25/67 (37%), Positives = 34/67 (50%)
Frame = -3
Query: 777 AAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQN 836
A G G + GR ++ +Q+R G G G GG +N+ GG G G G GGGGY++
Sbjct: 459 AIGAMNGQNLDGRNITVNEAQSRGGGGGGGGGG----YNRGGGGYGGRSGGGGGGGGYRS 292
Query: 837 SSRTQGG 843
GG
Sbjct: 291 RDGGYGG 271
>TC214360 similar to UP|Q9MBF3 (Q9MBF3) Glycine-rich RNA-binding protein,
partial (44%)
Length = 630
Score = 49.3 bits (116), Expect = 7e-06
Identities = 36/88 (40%), Positives = 37/88 (41%)
Frame = +2
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG R G Y GGGGG GG R G G GYGG +
Sbjct: 2 GGGGYS-----RGGGGY-----GGGGGRREGG--------YNRNGGG-GGYGGGGGGYGG 124
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGS 844
GG GG R RG GG G SR GGS
Sbjct: 125 GGGYGGGGRDRGYGGDGGSRYSRGGGGS 208
Score = 29.6 bits (65), Expect = 6.0
Identities = 25/72 (34%), Positives = 26/72 (35%)
Frame = +2
Query: 780 GGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSR 839
GGGG S GG G G GGR GGY+R GGGGY
Sbjct: 2 GGGGYSRGG-------------GGYGGGGGRR--------EGGYNRNG--GGGGYGGGGG 112
Query: 840 TQGGSALRGPHG 851
GG G G
Sbjct: 113 GYGGGGGYGGGG 148
>TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein,
complete
Length = 748
Score = 48.5 bits (114), Expect = 1e-05
Identities = 30/87 (34%), Positives = 38/87 (43%)
Frame = +1
Query: 747 SNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAG 806
S + +A G G +L R + + GGGGG GG Y R G G
Sbjct: 238 SEQSMKDAIEGMNGQNLDGRNITVNEAQSRGGGGGGGGGGGG------YGGGRGGGYGGG 399
Query: 807 YGGRALSFNQAGGSGGYSRGRGMGGGG 833
G +N++GG GGY G G GGGG
Sbjct: 400 GRGGGGGYNRSGGGGGYGGGGGYGGGG 480
Score = 47.0 bits (110), Expect = 4e-05
Identities = 34/94 (36%), Positives = 41/94 (43%), Gaps = 2/94 (2%)
Frame = +1
Query: 752 SEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRA 811
+++R GGGG G G GGG GGR Y+++ G G G GG
Sbjct: 316 AQSRGGGGG--------GGGGGGGYGGGRGGGYGGGGRGGGGGYNRSGGGGGYGGGGGYG 471
Query: 812 LSFNQAGGSGGYSRGR--GMGGGGYQNSSRTQGG 843
GG GGY GR G GGGG + SR G
Sbjct: 472 -----GGGGGGYGGGRDRGYGGGGDRGYSRGGDG 558
Score = 46.6 bits (109), Expect = 5e-05
Identities = 32/91 (35%), Positives = 36/91 (39%)
Frame = +1
Query: 746 ESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRA 805
E+ R GGGG R G Y GGGGG + G G G
Sbjct: 313 EAQSRGGGGGGGGGGGGYGGGRGGG--YGGGGRGGGGGYNRSGGGGGYGGGGGYGGGGGG 486
Query: 806 GYGGRALSFNQAGGSGGYSRGRGMGGGGYQN 836
GYGG GG GYSRG G GG++N
Sbjct: 487 GYGGGRDRGYGGGGDRGYSRGGDGGDGGWRN 579
>TC214476 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (14%)
Length = 642
Score = 47.8 bits (112), Expect = 2e-05
Identities = 29/83 (34%), Positives = 34/83 (40%)
Frame = -3
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGM 829
G+ + G GGG S GG GSG GYGG ++ GGS G G G
Sbjct: 502 GEGSGSQGGGSGGGGSQGGG----------SGSGGGGYGGGGSGGSEGGGSSGGGGGGGG 353
Query: 830 GGGGYQNSSRTQGGSALRGPHGD 852
GGGG G + G GD
Sbjct: 352 GGGGGGYGGGGDKGGGIGGDKGD 284
Score = 46.6 bits (109), Expect = 5e-05
Identities = 36/100 (36%), Positives = 46/100 (46%), Gaps = 5/100 (5%)
Frame = -3
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG + G Y + GGGG++ GG + GSG + GG S +Q
Sbjct: 634 GGGGSAGGGGSKGGGGYGGGGSQGGGGSAGGG--------GS*GGSGGSAGGGEG-SGSQ 482
Query: 817 AGGSGG---YSRGRGMGGGGY--QNSSRTQGGSALRGPHG 851
GGSGG G G GGGGY S ++GG + G G
Sbjct: 481 GGGSGGGGSQGGGSGSGGGGYGGGGSGGSEGGGSSGGGGG 362
>BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (25%)
Length = 407
Score = 47.0 bits (110), Expect = 4e-05
Identities = 32/94 (34%), Positives = 35/94 (37%)
Frame = +1
Query: 750 RASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGG 809
R E GGGG G+ GGGGG GG + R+G R G GG
Sbjct: 91 RGGEGGGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGGGGGGVF*GGERXREGGERGGGGG 270
Query: 810 RALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
GG GG G G GGGG GG
Sbjct: 271 GGGGGGGGGGXGGGGGGGGRGGGGGGGGGGGGGG 372
Score = 46.6 bits (109), Expect = 5e-05
Identities = 32/92 (34%), Positives = 33/92 (35%)
Frame = +3
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G GGGGG GG L + G G G GG
Sbjct: 138 GGGG---------GGRGGGGGGGGGGGGGGGGGGGFLGGREXXGGGGEGGGGGGGGGGGG 290
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRG 848
GG GG GRG GGGG GG G
Sbjct: 291 GGGXGGGGGGRGEGGGGGGGGGGGGGGGGEGG 386
Score = 43.9 bits (102), Expect = 3e-04
Identities = 28/77 (36%), Positives = 30/77 (38%)
Frame = +1
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGG R R+G + GGGGG GG G G GGR
Sbjct: 205 GGGVF*GGERXREGGERGGGGGGGGGGGGGGGX------------GGGGGGGGRGGGGGG 348
Query: 817 AGGSGGYSRGRGMGGGG 833
GG GG GR GGGG
Sbjct: 349 GGGGGGGGEGREEGGGG 399
Score = 43.1 bits (100), Expect = 5e-04
Identities = 30/95 (31%), Positives = 35/95 (36%)
Frame = +2
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG + G GGGGG G + + +G G G GG
Sbjct: 122 GGGGGGGGGGGKGGGGGGGGGGGGGGGGGGGFFRGERXXGRGGRGGGGGGGGGGGGGGGG 301
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
GG GG G G GGGG +GG G G
Sbjct: 302 GGGGGGEGGGGGGGGGGGGGGGGGRGGRREEGGGG 406
Score = 42.4 bits (98), Expect = 9e-04
Identities = 27/75 (36%), Positives = 31/75 (41%)
Frame = +1
Query: 759 GGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAG 818
GG ++ P G GGGGG GGR + G G G GG + G
Sbjct: 1 GGGEVCPGGGGGGGGGGGGGGGGGGGGGGGRGGE------GGGGGGGGGGGGGGGGGEGG 162
Query: 819 GSGGYSRGRGMGGGG 833
G GG G G GGGG
Sbjct: 163 GGGGGGGGGGGGGGG 207
Score = 42.0 bits (97), Expect = 0.001
Identities = 29/82 (35%), Positives = 32/82 (38%), Gaps = 3/82 (3%)
Frame = +2
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGR---ALSFNQAGGSGGYSRG 826
G+ +A GGGGG GG G G G GGR GG GG +G
Sbjct: 5 GERFARGGGGGGGGGGGGGGGGG--------GGGGGGGGGRGGGGGGGGGGGGGGGGGKG 160
Query: 827 RGMGGGGYQNSSRTQGGSALRG 848
G GGGG GG RG
Sbjct: 161 GGGGGGGGGGGGGGGGGGFFRG 226
Score = 40.0 bits (92), Expect = 0.004
Identities = 30/85 (35%), Positives = 30/85 (35%)
Frame = +3
Query: 759 GGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAG 818
GG LP G GGGGG GG G G G GG G
Sbjct: 3 GGRGLP-----GGGGGGGGGGGGGGGGGGG------------GGGGEGGGGGGGGGGGGG 131
Query: 819 GSGGYSRGRGMGGGGYQNSSRTQGG 843
G GG GRG GGGG GG
Sbjct: 132 GGGGGGGGRGGGGGGGGGGGGGGGG 206
Score = 39.7 bits (91), Expect = 0.006
Identities = 31/95 (32%), Positives = 32/95 (33%)
Frame = +2
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG R G GGGGG GG G G G GG +
Sbjct: 74 GGGGGGGGGRGGGGGGGGGGGGGGGGGKGGGG-------GGGGGGGGGGGGGGGFFRGER 232
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
G GG G G GGGG GG G G
Sbjct: 233 XXGRGGRGGGGGGGGGGGGGGGGGGGGGGEGGGGG 337
Score = 35.8 bits (81), Expect = 0.083
Identities = 26/73 (35%), Positives = 27/73 (36%)
Frame = +1
Query: 779 GGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSS 838
GGGGG GG G G G GG GG GG G G GGGG
Sbjct: 25 GGGGGGGGGG------------GGGGGGGGG-------GGGRGGEGGGGGGGGGGGGGGG 147
Query: 839 RTQGGSALRGPHG 851
+GG G G
Sbjct: 148 GGEGGGGGGGGGG 186
Score = 34.7 bits (78), Expect = 0.19
Identities = 27/82 (32%), Positives = 30/82 (35%)
Frame = +3
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGM 829
G+ GGGGG GG G G G GG + GG GG G G
Sbjct: 6 GRGLPGGGGGGGGGGGGGG------------GGGGGGGGG------EGGGGGG---GGGG 122
Query: 830 GGGGYQNSSRTQGGSALRGPHG 851
GGGG +GG G G
Sbjct: 123 GGGGGGGGGGGRGGGGGGGGGG 188
Score = 30.8 bits (68), Expect = 2.7
Identities = 18/44 (40%), Positives = 18/44 (40%)
Frame = +3
Query: 808 GGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
GGR L GG GG G G GGGG GG G G
Sbjct: 3 GGRGLPGGGGGGGGGGGGGGGGGGGGGGGEGGGGGGGGGGGGGG 134
>TC214511 weakly similar to PIR|C85356|C85356 glycine-rich protein [imported]
- Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(88%)
Length = 1085
Score = 45.4 bits (106), Expect = 1e-04
Identities = 28/65 (43%), Positives = 35/65 (53%), Gaps = 5/65 (7%)
Frame = +2
Query: 774 AAAAAGGGGGTSSGGRWQDLSYS---QTRQGSGRAGYGGRALSF--NQAGGSGGYSRGRG 828
A A AG G G+ SG S S + GSG +G G A S+ ++AG G S+GRG
Sbjct: 539 AGAGAGSGSGSGSGSSSSSASSSASSSSSSGSGGSGAGSEAGSYAGSRAGSGSGRSQGRG 718
Query: 829 MGGGG 833
GGGG
Sbjct: 719 GGGGG 733
Score = 33.9 bits (76), Expect = 0.32
Identities = 25/77 (32%), Positives = 31/77 (39%)
Frame = +2
Query: 758 GGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQA 817
G G +++ +GG G S G SY+ +R GSG GR
Sbjct: 572 GSGSSSSSASSSASSSSSSGSGGSGAGSEAG-----SYAGSRAGSGSGRSQGRG-----G 721
Query: 818 GGSGGYSRGRGMGGGGY 834
GG GG G G GG GY
Sbjct: 722 GGGGGGGGGGG-GGSGY 769
Score = 30.4 bits (67), Expect = 3.5
Identities = 21/67 (31%), Positives = 24/67 (35%)
Frame = +2
Query: 766 RRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSR 825
R G + GGGGG GG G G +GYG G G+
Sbjct: 680 RAGSGSGRSQGRGGGGGGGGGGG------------GGGGSGYG--------HGEGYGHGE 799
Query: 826 GRGMGGG 832
G G GGG
Sbjct: 800 GYGQGGG 820
>TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein-1, partial
(77%)
Length = 883
Score = 43.9 bits (102), Expect = 3e-04
Identities = 22/52 (42%), Positives = 25/52 (47%)
Frame = +3
Query: 800 QGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
QG+ R G GGR+ + GG GGY G G GGGG GG G G
Sbjct: 276 QGTRRGGDGGRSYGGGRGGGGGGYGGGGGYGGGGGYGGGGGYGGGGRGGGGG 431
Score = 40.4 bits (93), Expect = 0.003
Identities = 30/79 (37%), Positives = 32/79 (39%), Gaps = 2/79 (2%)
Frame = +3
Query: 769 DGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGY-GGRALSFNQAGGSGGYSRGR 827
DG+ A G G + G R R G G Y GGR GG GGY G
Sbjct: 225 DGRAKAVDVTGPDGASVQGTR---------RGGDGGRSYGGGRGGGGGGYGGGGGYGGGG 377
Query: 828 GM-GGGGYQNSSRTQGGSA 845
G GGGGY R GG A
Sbjct: 378 GYGGGGGYGGGGRGGGGGA 434
Score = 40.4 bits (93), Expect = 0.003
Identities = 36/119 (30%), Positives = 50/119 (41%), Gaps = 15/119 (12%)
Frame = +3
Query: 731 QALAFQLTEKLSILAESNERASEARLGG-GGLDLPPRRRDG---QDYAAAAAGGGGGTSS 786
++LA + + +I +ES+ RA + G G + RR G + Y GGGGG
Sbjct: 174 RSLAEGESVEFAIESESDGRAKAVDVTGPDGASVQGTRRGGDGGRSYGGGRGGGGGGYGG 353
Query: 787 GGRWQD---LSYSQTRQGSGRAGYGGRALSFNQAG--------GSGGYSRGRGMGGGGY 834
GG + G GR G GG + ++G G GG G G GGG Y
Sbjct: 354 GGGYGGGGGYGGGGGYGGGGRGGGGGACYNCGESGHLARDCSGGGGGDRYGGGGGGGRY 530
Score = 38.5 bits (88), Expect = 0.013
Identities = 30/82 (36%), Positives = 31/82 (37%), Gaps = 5/82 (6%)
Frame = +3
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGT-----SSGGRWQDLSYSQTRQGSGRAGYGGRA 811
GGGG G Y GGGGG SG +D S G G YGG
Sbjct: 351 GGGGYGGGGGYGGGGGYGGGGRGGGGGACYNCGESGHLARDCS-----GGGGGDRYGGGG 515
Query: 812 LSFNQAGGSGGYSRGRGMGGGG 833
GG GG G G GGGG
Sbjct: 516 GGGRYGGGGGGRYVGGGGGGGG 581
>TC225230 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (Fibrillarin
2) (AtFib2), partial (66%)
Length = 773
Score = 43.9 bits (102), Expect = 3e-04
Identities = 32/81 (39%), Positives = 35/81 (42%)
Frame = +1
Query: 764 PPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGY 823
PPR R G GGGGG GGR + R G GR G G + GG GG
Sbjct: 31 PPRGRGG-------FGGGGGFRGGGRGDR---GRGRGGGGRGGDRGTPFKA-RGGGRGGG 177
Query: 824 SRGRGMGGGGYQNSSRTQGGS 844
RG G GGG +GGS
Sbjct: 178 GRGGGRGGGRGGGRGGMKGGS 240
>TC214257 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (7%)
Length = 607
Score = 43.5 bits (101), Expect = 4e-04
Identities = 31/78 (39%), Positives = 35/78 (44%), Gaps = 1/78 (1%)
Frame = +2
Query: 780 GGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSR 839
GGGG GGR R+G G GG +N+ GG GGY G G GGGG
Sbjct: 8 GGGGYGGGGR---------REGGG----GG----YNRNGGGGGYGGGGGYGGGGGHGGGG 136
Query: 840 TQGGSAL-RGPHGDVSTR 856
GG RG GD +R
Sbjct: 137 GYGGGGRDRGYGGDGGSR 190
Score = 39.3 bits (90), Expect = 0.008
Identities = 30/77 (38%), Positives = 33/77 (41%)
Frame = +2
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG RR+G GGGGG GG + G G G GGR +
Sbjct: 23 GGGG------RREGGGGGYNRNGGGGGYGGGGGYGG---GGGHGGGGGYGGGGRDRGYGG 175
Query: 817 AGGSGGYSRGRGMGGGG 833
GGS YSRG G GG
Sbjct: 176 DGGS-RYSRGGGASDGG 223
Score = 38.5 bits (88), Expect = 0.013
Identities = 31/88 (35%), Positives = 32/88 (36%)
Frame = +2
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG RR G GGGG GG G G+GG
Sbjct: 8 GGGGYGGGGRREGGGGGYNRNGGGGGYGGGGGY------------GGGGGHGG------- 130
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGS 844
GG GG R RG GG G SR G S
Sbjct: 131 GGGYGGGGRDRGYGGDGGSRYSRGGGAS 214
>BE661383 similar to GP|8778710|gb| T1N15.2 {Arabidopsis thaliana}, partial
(6%)
Length = 591
Score = 42.0 bits (97), Expect = 0.001
Identities = 41/126 (32%), Positives = 50/126 (39%), Gaps = 17/126 (13%)
Frame = +1
Query: 736 QLTEKLSILAESNERASEARLGGGGLDLP-PRRRDG---QDYAAAAAGGGGGTSSGGRWQ 791
Q+ ++ +R +E GG + P R Q AAAAA GG G G W
Sbjct: 76 QMCPMRAVFVMVRKRRTELPSGGESSEAQHPSERSAPPPQQQAAAAAPGGAGPQGGRGWG 255
Query: 792 DLSYSQTRQGSGRA------------GYGGRALSF-NQAGGSGGYSRGRGMGGGGYQNSS 838
Q +G GR+ Y GR +Q GG GGY GR GGGG S
Sbjct: 256 P----QGGRGGGRSRGMPQQQYGAPPDYQGRGRGGPSQQGGRGGYGSGRS-GGGGGMGSG 420
Query: 839 RTQGGS 844
R G S
Sbjct: 421 RGVGPS 438
>TC211031 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (11%)
Length = 678
Score = 42.0 bits (97), Expect = 0.001
Identities = 31/89 (34%), Positives = 35/89 (38%)
Frame = -3
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG R G + GGG G GG D + G G G GG N
Sbjct: 403 GGGGASGGGDGRFGGEGDGGKGGGGDGGKGGG--VDSGIGGGKGGGGDGGNGGGGDGGNG 230
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSA 845
GG GG G G GG +S GGS+
Sbjct: 229 GGGDGGNGGGGASGIGGGGHSLGGGGGSS 143
Score = 39.7 bits (91), Expect = 0.006
Identities = 29/96 (30%), Positives = 35/96 (36%)
Frame = -3
Query: 756 LGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFN 815
+G GG G A+ GGGG S GG + +G G G G +
Sbjct: 451 IGNGG--------SGGGGGASGGNGGGGASGGGDGRFGGEGDGGKGGGGDGGKGGGVDSG 296
Query: 816 QAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
GG GG G GGG N GG+ G G
Sbjct: 295 IGGGKGGGGDGGNGGGGDGGNGGGGDGGNGGGGASG 188
Score = 35.8 bits (81), Expect = 0.083
Identities = 28/95 (29%), Positives = 31/95 (32%)
Frame = -3
Query: 758 GGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQA 817
GGG P G + GGG +GG G G G GG
Sbjct: 490 GGGTRYSPGPFGGIGNGGSGGGGGASGGNGGGGASGGGDGRFGGEGDGGKGGG----GDG 323
Query: 818 GGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGD 852
G GG G G G GG + GG G GD
Sbjct: 322 GKGGGVDSGIGGGKGGGGDGGNGGGGDGGNGGGGD 218
>TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (GRP2)
(AT4g38680/F20M13_240), partial (42%)
Length = 891
Score = 41.6 bits (96), Expect = 0.002
Identities = 28/70 (40%), Positives = 29/70 (41%), Gaps = 2/70 (2%)
Frame = +3
Query: 780 GGGGTSSGGRWQDLSYSQTRQGSGRAGYGG--RALSFNQAGGSGGYSRGRGMGGGGYQNS 837
GGGG G SY G GR GYGG R GG GGY G GGGGY
Sbjct: 57 GGGGXXRG------SYGX---GGGRGGYGGGGRGGRGGYGGGGGGYGGGGYGGGGGYGGG 209
Query: 838 SRTQGGSALR 847
GG +
Sbjct: 210 GGGGGGGCYK 239
Score = 41.2 bits (95), Expect = 0.002
Identities = 26/62 (41%), Positives = 26/62 (41%)
Frame = +3
Query: 779 GGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSS 838
GG GG GGR Y G G GYGG GG GGY G G GGGG
Sbjct: 96 GGRGGYGGGGRGGRGGY-----GGGGGGYGGGGY-----GGGGGYGGGGGGGGGGCYKCG 245
Query: 839 RT 840
T
Sbjct: 246 ET 251
Score = 32.7 bits (73), Expect = 0.71
Identities = 18/48 (37%), Positives = 19/48 (39%)
Frame = +3
Query: 804 RAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
R G G S+ GG GGY G G GGY GG G G
Sbjct: 54 RGGGGXXRGSYGXGGGRGGYGGGGRGGRGGYGGGGGGYGGGGYGGGGG 197
Score = 32.3 bits (72), Expect = 0.92
Identities = 23/75 (30%), Positives = 27/75 (35%)
Frame = +3
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G Y GGGGG G ++ ++ G G YGG
Sbjct: 168 GGGGYG------GGGGYGGGGGGGGGGCYKCGETGHIARDCSQGGGGGGRYGG------- 308
Query: 817 AGGSGGYSRGRGMGG 831
GG GG G G
Sbjct: 309 GGGGGGSCYNCGESG 353
>BE661382 similar to GP|8778710|gb| T1N15.2 {Arabidopsis thaliana}, partial
(5%)
Length = 598
Score = 41.6 bits (96), Expect = 0.002
Identities = 37/115 (32%), Positives = 46/115 (39%), Gaps = 17/115 (14%)
Frame = +1
Query: 736 QLTEKLSILAESNERASEARLGGGGLDLP-PRRRDG---QDYAAAAAGGGGGTSSGGRWQ 791
Q+ ++ +R +E GG + P R Q AAAAA GG G G W
Sbjct: 76 QMCPMRAVFVMVRKRRTELPSGGESSEAQHPSERSAPPPQQQAAAAAPGGAGPQGGRGWG 255
Query: 792 DLSYSQTRQGSGRA------------GYGGRALSF-NQAGGSGGYSRGRGMGGGG 833
Q +G GR+ Y GR +Q GG GGY GR GGGG
Sbjct: 256 P----QGGRGGGRSRGMPQQQYGAPPDYQGRGRGGPSQQGGRGGYGSGRSGGGGG 408
>TC219649 weakly similar to UP|Q95JD1 (Q95JD1) Basic proline-rich protein,
partial (37%)
Length = 750
Score = 41.2 bits (95), Expect = 0.002
Identities = 33/108 (30%), Positives = 37/108 (33%)
Frame = -1
Query: 749 ERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYG 808
E S A G GG P G G GGG GG + G G YG
Sbjct: 414 EPGSGAGPGSGGYGGGP----GGGVGPGGGGPGGGAGRGGGGPGSGAGPSGGGPGGGSYG 247
Query: 809 GRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGDVSTR 856
G GG GG G G GGG + + G GP+G R
Sbjct: 246 GEP----GGGGPGGDGYGGGPGGGAGPDGGGPKDGGFGGGPYGGYGPR 115
Score = 35.4 bits (80), Expect = 0.11
Identities = 30/99 (30%), Positives = 36/99 (36%)
Frame = -1
Query: 753 EARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRAL 812
+A GGGG G+ + A G GG G G+GR G G +
Sbjct: 468 DAGPGGGGPGGGAGPGGGEPGSGAGPGSGGYGGGPGGGVGPGGGGPGGGAGRGGGGPGSG 289
Query: 813 SFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
+ GG GG S G GGGG G GP G
Sbjct: 288 AGPSGGGPGGGSYGGEPGGGGPGGDGYGGGPGGGAGPDG 172
Score = 35.0 bits (79), Expect = 0.14
Identities = 29/79 (36%), Positives = 32/79 (39%), Gaps = 4/79 (5%)
Frame = -1
Query: 778 AGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRG----RGMGGGG 833
AG GGG+ GG D G G G GG A GSGGY G G GGG
Sbjct: 666 AGSGGGSYGGGPGGDA-------GPGGGGPGGSA-----GPGSGGYGGGPGGDTGPGGGE 523
Query: 834 YQNSSRTQGGSALRGPHGD 852
+ + G GP GD
Sbjct: 522 PGSGAGPGSGGYGGGPGGD 466
Score = 31.6 bits (70), Expect = 1.6
Identities = 31/91 (34%), Positives = 33/91 (36%), Gaps = 4/91 (4%)
Frame = -1
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GG G D P G +AG G G GG D T G G G G
Sbjct: 639 GGPGGDAGP----GGGGPGGSAGPGSGGYGGGPGGD-----TGPGGGEPGSGA------- 508
Query: 817 AGGSGGYSRG----RGMGGGGYQNSSRTQGG 843
GSGGY G G GGGG + GG
Sbjct: 507 GPGSGGYGGGPGGDAGPGGGGPGGGAGPGGG 415
>TC205204 homologue to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (44%)
Length = 996
Score = 40.8 bits (94), Expect = 0.003
Identities = 37/120 (30%), Positives = 47/120 (38%), Gaps = 2/120 (1%)
Frame = -2
Query: 736 QLTEKLSILAESNERASEARLGGGGL-DLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLS 794
++ E+L A+ E A L G G ++ G A GGG + GG
Sbjct: 650 EMVERLGPGAKEGESAGATTLAGAGAGEVAGGEERGVGVNAGGGVAGGGVAGGGEKGGGE 471
Query: 795 YSQTRQGSGRAGYGGRALSFNQAGG-SGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGDV 853
G G AG GG A + GG +GG G G GGG + GG L G G V
Sbjct: 470 AGGGVAGGGDAG-GGEAGGGDAGGGDAGGGEAGGGDAGGGDEGGGEA-GGGELSGAGGGV 297
>TC214303 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, partial (18%)
Length = 1014
Score = 40.8 bits (94), Expect = 0.003
Identities = 30/84 (35%), Positives = 34/84 (39%), Gaps = 6/84 (7%)
Frame = -1
Query: 756 LGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQ------TRQGSGRAGYGG 809
+GGGG DG GGGGG GG W + QG G G GG
Sbjct: 375 VGGGG--------DGGGGELY*IGGGGGDGDGGGW*GAECGDGGGGECSSQGGGGGGGGG 220
Query: 810 RALSFNQAGGSGGYSRGRGMGGGG 833
++ Q GG GG G GGGG
Sbjct: 219 DSI---QGGGDGGGGGGE*TGGGG 157
Score = 35.0 bits (79), Expect = 0.14
Identities = 35/125 (28%), Positives = 44/125 (35%), Gaps = 16/125 (12%)
Frame = -1
Query: 744 LAESNERASEARLGGGG--LDLPPRRRDGQDYAAAAAG--------------GGGGTSSG 787
L E + + ++ + GGGG + P G Y A G GGGG G
Sbjct: 528 LEEKSSQKNQ*KGGGGGDAYETPMTGGKGPSYTGAGGGDDGGGGEL***TGVGGGGDGGG 349
Query: 788 GRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALR 847
G + G G G GG G GG +G GGGG QGG
Sbjct: 348 GELY*IG------GGGGDGDGGGW*GAECGDGGGGECSSQGGGGGG-GGGDSIQGGGDGG 190
Query: 848 GPHGD 852
G G+
Sbjct: 189 GGGGE 175
Score = 33.1 bits (74), Expect = 0.54
Identities = 27/75 (36%), Positives = 28/75 (37%)
Frame = -1
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG D DG GGGG + GG D G G G GG
Sbjct: 234 GGGGGDSIQGGGDGGGGGGE*TGGGGE*TGGGGGGDK*G*TGGGGGGECGGGG------D 73
Query: 817 AGGSGGYSRGRGMGG 831
GG GG G G GG
Sbjct: 72 GGGGGGEYVGGGAGG 28
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.134 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,066,391
Number of Sequences: 63676
Number of extensions: 390770
Number of successful extensions: 4839
Number of sequences better than 10.0: 265
Number of HSP's better than 10.0 without gapping: 3557
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4217
length of query: 865
length of database: 12,639,632
effective HSP length: 105
effective length of query: 760
effective length of database: 5,953,652
effective search space: 4524775520
effective search space used: 4524775520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC137602.5


