

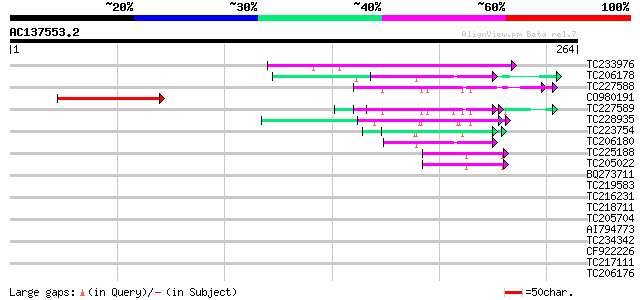
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137553.2 - phase: 0
(264 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC233976 51 6e-07
TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 51 6e-07
TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L1... 50 1e-06
CO980191 46 2e-05
TC227589 45 3e-05
TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana geno... 45 3e-05
TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequen... 45 4e-05
TC206180 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 43 1e-04
TC225188 similar to UP|Q9FYB7 (Q9FYB7) Splicing factor-like prot... 42 2e-04
TC205022 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, pa... 42 2e-04
BQ273711 37 0.007
TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich p... 36 0.015
TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (G... 35 0.035
TC218711 similar to UP|Q90Z60 (Q90Z60) Rev-Erb beta (Fragment), ... 35 0.035
TC205704 similar to UP|Q8W042 (Q8W042) Prenylated Rab receptor 2... 31 0.50
AI794773 similar to GP|11875626|gb XRN4 {Arabidopsis thaliana}, ... 31 0.50
TC234342 similar to UP|COLE_ARATH (Q9LUA9) Zinc finger protein c... 31 0.50
CF922226 31 0.50
TC217111 similar to UP|P93188 (P93188) NADPH-dependent HC-toxin ... 30 1.4
TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein... 28 3.2
>TC233976
Length = 763
Score = 50.8 bits (120), Expect = 6e-07
Identities = 32/125 (25%), Positives = 51/125 (40%), Gaps = 9/125 (7%)
Frame = +1
Query: 121 EEDTKAHYKIRNERRNKGQQ----GRPKPYSAPVNK-----GEQRLNDERRPKRRDDPAE 171
+++ A YK N K ++ R KPYSAP+ G++
Sbjct: 25 QQEKVAFYKNANASHGKEKKPMTHSRAKPYSAPLEYENHYGGQRTSGGHHLAGGSSQLVN 204
Query: 172 IVCYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPK 231
V G G + +C +CG+ H EC ++ CFN +GHL++ C P+
Sbjct: 205 RVSQPAGRGGSGAPAIVTTPLRCRKCGRLGHNAHECTYREVTCFNYQGKGHLSTNCPHPR 384
Query: 232 KGQAS 236
K + S
Sbjct: 385 KEKRS 399
>TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (69%)
Length = 1138
Score = 50.8 bits (120), Expect = 6e-07
Identities = 34/137 (24%), Positives = 54/137 (38%), Gaps = 2/137 (1%)
Frame = +2
Query: 123 DTKAHYKIRNERRNKGQQGRPKPYSAPVNKGEQRLNDE--RRPKRRDDPAEIVCYKCGEK 180
+ K +K+ ++ R++ + P + D RR RR + +C C
Sbjct: 185 ERKKKWKMSSDSRSRSRSRSRSPMDRKIRSDRFSYRDAPYRRDSRRGFSRDNLCKNCKRP 364
Query: 181 GHKSNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKKGQASVYDS 240
GH + C C CG H +EC + C+NC E GH+ S C P +G
Sbjct: 365 GHYARECPNVAI-CHNCGLPGHIASECTTKSL-CWNCKEPGHMASSC--PNEG------- 511
Query: 241 DYVLTKVMNKCGELGIR 257
+ + CG+ G R
Sbjct: 512 ------ICHTCGKAGHR 544
Score = 50.1 bits (118), Expect = 1e-06
Identities = 23/65 (35%), Positives = 34/65 (51%), Gaps = 6/65 (9%)
Frame = +2
Query: 169 PAEIVCYKCGEKGHKSNVCN------GEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGH 222
P E +C+ CG+ GH++ C+ G+ + C C K+ H AEC + C NC + GH
Sbjct: 500 PNEGICHTCGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAEC-TNEKACNNCRKTGH 676
Query: 223 LTSQC 227
L C
Sbjct: 677 LARDC 691
Score = 37.4 bits (85), Expect = 0.007
Identities = 15/40 (37%), Positives = 19/40 (47%)
Frame = +2
Query: 171 EIVCYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRG 210
++VC C + GH S C G C CG + H EC G
Sbjct: 833 DVVCRNCQQLGHMSRDCMGPLMICHNCGGRGHLAYECPSG 952
Score = 32.7 bits (73), Expect = 0.17
Identities = 19/87 (21%), Positives = 28/87 (31%), Gaps = 25/87 (28%)
Frame = +2
Query: 166 RDDPAEIVCYKCGEKGHKSNVC-------------------------NGEEKKCFRCGKK 200
RD P + +C C GH + C + C C +
Sbjct: 683 RDCPNDPICNLCNVSGHVARQCPKANVLGDRSGGGGGGGGARGGGGGGYRDVVCRNCQQL 862
Query: 201 WHTIAECKRGDIVCFNCDEEGHLTSQC 227
H +C ++C NC GHL +C
Sbjct: 863 GHMSRDCMGPLMICHNCGGRGHLAYEC 943
Score = 32.3 bits (72), Expect = 0.22
Identities = 19/85 (22%), Positives = 28/85 (32%), Gaps = 25/85 (29%)
Frame = +2
Query: 171 EIVCYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRG-------------------- 210
E C C + GH + C + C C H +C +
Sbjct: 641 EKACNNCRKTGHLARDCPNDPI-CNLCNVSGHVARQCPKANVLGDRSGGGGGGGGARGGG 817
Query: 211 -----DIVCFNCDEEGHLTSQCKKP 230
D+VC NC + GH++ C P
Sbjct: 818 GGGYRDVVCRNCQQLGHMSRDCMGP 892
>TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L16.11 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(10%)
Length = 1300
Score = 49.7 bits (117), Expect = 1e-06
Identities = 32/103 (31%), Positives = 42/103 (40%), Gaps = 13/103 (12%)
Frame = +1
Query: 161 RRPKRRDDPAEIVCYKCGEKGHKSNVCNGE----EKKCFRCGKKWHTIAECKR--GDIV- 213
R +DD EI CY C GH V + E C++CG+ HT C R +I
Sbjct: 307 RNDYSQDDLKEIQCYVCKRLGHLCCVNTDDATPGEISCYKCGQLGHTGLACSRLRDEITS 486
Query: 214 ------CFNCDEEGHLTSQCKKPKKGQASVYDSDYVLTKVMNK 250
CF C EEGH +C K ++S + K K
Sbjct: 487 GATPSSCFKCGEEGHFARECTSSIKSGKRNWESSHTKDKRSQK 615
Score = 45.8 bits (107), Expect = 2e-05
Identities = 30/99 (30%), Positives = 44/99 (44%), Gaps = 4/99 (4%)
Frame = +1
Query: 161 RRPKRRDDPAEI--VCYKCGEKGHKSNVCNGEEKK--CFRCGKKWHTIAECKRGDIVCFN 216
R P+ D P C+ CGE+GH + C+ ++K C+ CG H +C + CF
Sbjct: 16 RGPRYFDPPDNSWGACFNCGEEGHAAVNCSAVKRKKPCYVCGCLGHNARQCSKVQ-DCFI 192
Query: 217 CDEEGHLTSQCKKPKKGQASVYDSDYVLTKVMNKCGELG 255
C + GH C P+K + S + KCG G
Sbjct: 193 CKKGGHRAKDC--PEK-----HTSTSKSIAICLKCGNSG 288
>CO980191
Length = 802
Score = 45.8 bits (107), Expect = 2e-05
Identities = 23/50 (46%), Positives = 30/50 (60%)
Frame = +1
Query: 23 VVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVTEYAAKFVELAK 72
V+T VF+ FL ++ ED+ K+ EFLELK G+M V Y F EL K
Sbjct: 361 VIT*EVFKEIFLEKHLHEDI*NHKKTEFLELKHGNMIVANYLVNFDELPK 510
>TC227589
Length = 547
Score = 45.4 bits (106), Expect = 3e-05
Identities = 23/71 (32%), Positives = 33/71 (46%), Gaps = 4/71 (5%)
Frame = +2
Query: 161 RRPKRRDDPAEI--VCYKCGEKGHKSNVCNGEEKK--CFRCGKKWHTIAECKRGDIVCFN 216
R P+ D P C+ CGE GH + C+ ++K C+ CG H +C + CF
Sbjct: 2 RGPRYFDPPDSSWGACFNCGEDGHAAVNCSAAKRKKPCYVCGGLGHNARQCTKAQ-DCFI 178
Query: 217 CDEEGHLTSQC 227
C + GH C
Sbjct: 179 CKKGGHRAKDC 211
Score = 42.7 bits (99), Expect = 2e-04
Identities = 25/77 (32%), Positives = 32/77 (41%), Gaps = 13/77 (16%)
Frame = +2
Query: 167 DDPAEIVCYKCGEKGHKSNVCNGE----EKKCFRCGKKWHTIAECKR--GDIV------- 213
DD EI CY C GH V + E C++CG+ HT C + +I
Sbjct: 311 DDLKEIQCYVCKRVGHLCCVNTDDATPGEISCYKCGQLGHTGLACSKLPDEITSAATPSS 490
Query: 214 CFNCDEEGHLTSQCKKP 230
C C E GH +C P
Sbjct: 491 CCKCGEAGHFAQECTSP 541
Score = 41.6 bits (96), Expect = 4e-04
Identities = 30/116 (25%), Positives = 43/116 (36%), Gaps = 12/116 (10%)
Frame = +2
Query: 152 KGEQRLNDERRPKRRDDPAEIVCYKCGEKGHKSNVCNGE-------EKKCFRCGKKWHTI 204
KG R D + +C KCG GH C + E +C+ C + H
Sbjct: 185 KGGHRAKDCLEKHTSRSKSVAICLKCGNSGHDMFSCRNDYSPDDLKEIQCYVCKRVGHLC 364
Query: 205 A----ECKRGDIVCFNCDEEGHLTSQCKK-PKKGQASVYDSDYVLTKVMNKCGELG 255
+ G+I C+ C + GH C K P + ++ S KCGE G
Sbjct: 365 CVNTDDATPGEISCYKCGQLGHTGLACSKLPDEITSAATPSSCC------KCGEAG 514
Score = 28.1 bits (61), Expect = 4.2
Identities = 9/19 (47%), Positives = 11/19 (57%)
Frame = +2
Query: 214 CFNCDEEGHLTSQCKKPKK 232
CFNC E+GH C K+
Sbjct: 47 CFNCGEDGHAAVNCSAAKR 103
>TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K24M7, partial (36%)
Length = 1224
Score = 45.1 bits (105), Expect = 3e-05
Identities = 21/84 (25%), Positives = 39/84 (46%), Gaps = 13/84 (15%)
Frame = +1
Query: 163 PKRRDDPAEIVCYKCGEKGHKSNVCNG------EEKKCFRCGKKWHTIAEC----KRGDI 212
P++ + +C +C +GH++ C + K C+ CG+ H + +C + G
Sbjct: 313 PEKAEWEKNKICLRCRRRGHRAKNCPEVLDGAKDAKYCYNCGENGHALTQCLHPLQEGGT 492
Query: 213 V---CFNCDEEGHLTSQCKKPKKG 233
CF C++ GHL+ C + G
Sbjct: 493 KFAECFVCNQRGHLSKNCPQNTHG 564
Score = 40.8 bits (94), Expect = 6e-04
Identities = 31/140 (22%), Positives = 51/140 (36%), Gaps = 27/140 (19%)
Frame = +1
Query: 118 MIYEEDTKAHYKIRNERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDD--------- 168
M+ + A + ++E + +P P P K ++ N + KR +
Sbjct: 58 MVSQRQRLARKRYKSEHPELFPKPKPTPPKDPDKKKKKNKNSAFKRKRPEPKPGSRKRHL 237
Query: 169 -------PAEIVCYKCGEKGHKSNVCNGE-----EKKCFRCGKKWHTIAECK------RG 210
P E C+ C H + +C + K C RC ++ H C +
Sbjct: 238 LRVPGMKPGES-CFICKAMDHIAKLCPEKAEWEKNKICLRCRRRGHRAKNCPEVLDGAKD 414
Query: 211 DIVCFNCDEEGHLTSQCKKP 230
C+NC E GH +QC P
Sbjct: 415 AKYCYNCGENGHALTQCLHP 474
>TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequence, partial
(22%)
Length = 742
Score = 44.7 bits (104), Expect = 4e-05
Identities = 27/89 (30%), Positives = 32/89 (35%), Gaps = 22/89 (24%)
Frame = +2
Query: 165 RRDDPAEIVCYKCGEKGHKSNVCN----------GEEKKCFRCGKKWHTIAECKR----- 209
RR+ CY+CGE GH + CN G CF CG H +C R
Sbjct: 17 RRNGGGGAACYQCGEFGHLARDCNRSSNSGGGGGGSGGGCFNCGGFGHLARDCVRGGGGS 196
Query: 210 -------GDIVCFNCDEEGHLTSQCKKPK 231
G CF C GH+ C K
Sbjct: 197 VGIGGGGGGGSCFRCGGFGHMARDCATGK 283
Score = 42.7 bits (99), Expect = 2e-04
Identities = 22/78 (28%), Positives = 29/78 (36%), Gaps = 24/78 (30%)
Frame = +2
Query: 174 CYKCGEKGHKSNVC---------NGEEKKCFRCGKKWHTIAECKR--------------- 209
C++CG GH + C G CFRCG+ H +C
Sbjct: 230 CFRCGGFGHMARDCATGKGNIGGGGSGGGCFRCGEVGHLARDCGMEGGRFGGGGGSGGGG 409
Query: 210 GDIVCFNCDEEGHLTSQC 227
G CFNC + GH +C
Sbjct: 410 GKSTCFNCGKPGHFAREC 463
>TC206180 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (42%)
Length = 655
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/59 (33%), Positives = 30/59 (49%), Gaps = 6/59 (10%)
Frame = +3
Query: 175 YKCGEKGHKSNVCN------GEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQC 227
+ CG+ GH++ C+ G+ + C C K+ H AEC + C NC + GHL C
Sbjct: 3 HTCGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAEC-TNEKACNNCRKTGHLARDC 176
Score = 37.4 bits (85), Expect = 0.007
Identities = 15/40 (37%), Positives = 19/40 (47%)
Frame = +3
Query: 171 EIVCYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRG 210
++VC C + GH S C G C CG + H EC G
Sbjct: 309 DVVCRNCQQLGHMSRDCMGPLMICHNCGGRGHLAYECPSG 428
Score = 33.9 bits (76), Expect = 0.077
Identities = 19/84 (22%), Positives = 28/84 (32%), Gaps = 22/84 (26%)
Frame = +3
Query: 166 RDDPAEIVCYKCGEKGHKSNVC----------------------NGEEKKCFRCGKKWHT 203
RD P + +C C GH + C + C C + H
Sbjct: 168 RDCPNDPICNLCNVSGHVARQCPKANVLGDXXGGGGGARGGGGGGYRDVVCRNCQQLGHM 347
Query: 204 IAECKRGDIVCFNCDEEGHLTSQC 227
+C ++C NC GHL +C
Sbjct: 348 SRDCMGPLMICHNCGGRGHLAYEC 419
Score = 33.5 bits (75), Expect = 0.10
Identities = 19/82 (23%), Positives = 28/82 (33%), Gaps = 22/82 (26%)
Frame = +3
Query: 171 EIVCYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRG-------------------- 210
E C C + GH + C + C C H +C +
Sbjct: 126 EKACNNCRKTGHLARDCPNDPI-CNLCNVSGHVARQCPKANVLGDXXGGGGGARGGGGGG 302
Query: 211 --DIVCFNCDEEGHLTSQCKKP 230
D+VC NC + GH++ C P
Sbjct: 303 YRDVVCRNCQQLGHMSRDCMGP 368
>TC225188 similar to UP|Q9FYB7 (Q9FYB7) Splicing factor-like protein, partial
(62%)
Length = 1131
Score = 42.4 bits (98), Expect = 2e-04
Identities = 19/43 (44%), Positives = 23/43 (53%), Gaps = 3/43 (6%)
Frame = +1
Query: 193 KCFRCGKKWHTIAECKRGD--IVCFNCDEEGHLTSQCK-KPKK 232
+CF CG H +CK GD C+ C E GH+ CK PKK
Sbjct: 361 RCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCKNSPKK 489
Score = 37.7 bits (86), Expect = 0.005
Identities = 18/58 (31%), Positives = 25/58 (43%), Gaps = 2/58 (3%)
Frame = +1
Query: 153 GEQRLNDERRPKRRDDPAEIVCYKCGEKGHKSNVCNGEE--KKCFRCGKKWHTIAECK 208
G + D R P C+ CG GH + C + KC+RCG++ H CK
Sbjct: 301 GPRGSRDREYMGRGPPPGSGRCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCK 474
Score = 32.0 bits (71), Expect = 0.29
Identities = 11/27 (40%), Positives = 16/27 (58%)
Frame = +1
Query: 174 CYKCGEKGHKSNVCNGEEKKCFRCGKK 200
CY+CGE+GH C KK + G++
Sbjct: 430 CYRCGERGHIEKNCKNSPKKLSQRGRR 510
>TC205022 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, partial (69%)
Length = 1623
Score = 42.4 bits (98), Expect = 2e-04
Identities = 19/43 (44%), Positives = 23/43 (53%), Gaps = 3/43 (6%)
Frame = +2
Query: 193 KCFRCGKKWHTIAECKRGD--IVCFNCDEEGHLTSQCK-KPKK 232
+CF CG H +CK GD C+ C E GH+ CK PKK
Sbjct: 311 RCFNCGLDGHWARDCKAGDWKNKCYRCGERGHIERNCKNSPKK 439
Score = 37.0 bits (84), Expect = 0.009
Identities = 16/46 (34%), Positives = 22/46 (47%), Gaps = 2/46 (4%)
Frame = +2
Query: 165 RRDDPAEIVCYKCGEKGHKSNVCNGEE--KKCFRCGKKWHTIAECK 208
R P C+ CG GH + C + KC+RCG++ H CK
Sbjct: 287 RGPPPGSGRCFNCGLDGHWARDCKAGDWKNKCYRCGERGHIERNCK 424
Score = 30.0 bits (66), Expect = 1.1
Identities = 10/20 (50%), Positives = 12/20 (60%)
Frame = +2
Query: 174 CYKCGEKGHKSNVCNGEEKK 193
CY+CGE+GH C KK
Sbjct: 380 CYRCGERGHIERNCKNSPKK 439
>BQ273711
Length = 409
Score = 37.4 bits (85), Expect = 0.007
Identities = 15/37 (40%), Positives = 21/37 (56%)
Frame = -2
Query: 1 MLAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRY 37
ML EA++WWK + P G V+ W F+ +FL Y
Sbjct: 111 MLQGEAENWWKFVKPSFVAPGGVIPWNAFKDKFLENY 1
>TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich protein 2,
partial (45%)
Length = 995
Score = 36.2 bits (82), Expect = 0.015
Identities = 22/67 (32%), Positives = 27/67 (39%), Gaps = 7/67 (10%)
Frame = +2
Query: 174 CYKCGEKGHKSNVC-------NGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQ 226
CY CG GH + C G++ + R G G CFNC EEGH +
Sbjct: 425 CYNCGRIGHLARDCYHGQGGGGGDDGRNRRRGGGGG-------GGGGCFNCGEEGHFARE 583
Query: 227 CKKPKKG 233
C KG
Sbjct: 584 CPNVGKG 604
>TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (GRP2)
(AT4g38680/F20M13_240), partial (42%)
Length = 891
Score = 35.0 bits (79), Expect = 0.035
Identities = 18/54 (33%), Positives = 22/54 (40%)
Frame = +3
Query: 174 CYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQC 227
CYKCGE GH + + C + G G C+NC E GH C
Sbjct: 231 CYKCGETGHIA-------RDCSQGGGGGGRYGGGGGGGGSCYNCGESGHFARDC 371
>TC218711 similar to UP|Q90Z60 (Q90Z60) Rev-Erb beta (Fragment), partial (6%)
Length = 1204
Score = 35.0 bits (79), Expect = 0.035
Identities = 20/85 (23%), Positives = 34/85 (39%), Gaps = 12/85 (14%)
Frame = -3
Query: 161 RRPKRRDDPAEIVCYKCGEKGHKSNVCNG------EEKKCFRCGKKWHTIAECKRGD--- 211
+ P + + + C C + GH+ C +E+ CF CG+ H++ +C
Sbjct: 560 KSPFKHHGESSLRCRACRQPGHRFQQCQRLKCLSMDEEVCFFCGEIGHSLGKCDVSQAGG 381
Query: 212 ---IVCFNCDEEGHLTSQCKKPKKG 233
C C GH + C P+ G
Sbjct: 380 GRFAKCLLCYGHGHFSYNC--PQNG 312
Score = 30.8 bits (68), Expect = 0.65
Identities = 18/58 (31%), Positives = 22/58 (37%), Gaps = 6/58 (10%)
Frame = -3
Query: 171 EIVCYKCGEKGHKSNVCN------GEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGH 222
E VC+ CGE GH C+ G KC C H +NC + GH
Sbjct: 452 EEVCFFCGEIGHSLGKCDVSQAGGGRFAKCLLCYGHGH----------FSYNCPQNGH 309
>TC205704 similar to UP|Q8W042 (Q8W042) Prenylated Rab receptor 2, partial
(92%)
Length = 1038
Score = 31.2 bits (69), Expect = 0.50
Identities = 20/57 (35%), Positives = 27/57 (47%)
Frame = -1
Query: 134 RRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKGHKSNVCNGE 190
RR K + RP+ A NK +R+ DER +R D I+ + E VC GE
Sbjct: 528 RRAKEVKERPRREEAKENKERERMRDERDRQRERD*G*IIDAEVAEVLADPRVCFGE 358
>AI794773 similar to GP|11875626|gb XRN4 {Arabidopsis thaliana}, partial
(14%)
Length = 440
Score = 31.2 bits (69), Expect = 0.50
Identities = 10/20 (50%), Positives = 16/20 (80%)
Frame = +1
Query: 189 GEEKKCFRCGKKWHTIAECK 208
G+++KCF+CG+ H AEC+
Sbjct: 31 GQQEKCFQCGQLGHFAAECR 90
>TC234342 similar to UP|COLE_ARATH (Q9LUA9) Zinc finger protein constans-like
14, partial (27%)
Length = 688
Score = 31.2 bits (69), Expect = 0.50
Identities = 20/73 (27%), Positives = 29/73 (39%), Gaps = 3/73 (4%)
Frame = +3
Query: 166 RDDPAEIVCYKCGEKGHKSNVCNGEEKK---CFRCGKKWHTIAECKRGDIVCFNCDEEGH 222
R D A +C C H +N + + C RC + + + +C NCD GH
Sbjct: 342 RSDSA-CLCLSCDRNVHSANALSRRHSRTLVCERCNSQPAFVRSVEEKISLCQNCDWLGH 518
Query: 223 LTSQCKKPKKGQA 235
TS K Q+
Sbjct: 519 GTSPSSSMHKRQS 557
>CF922226
Length = 667
Score = 31.2 bits (69), Expect = 0.50
Identities = 21/84 (25%), Positives = 34/84 (40%), Gaps = 7/84 (8%)
Frame = -3
Query: 116 SYMIYEEDTKAHYKIRNERRNK-------GQQGRPKPYSAPVNKGEQRLNDERRPKRRDD 168
S + E T + K NER+ K G R K + +++ E + +
Sbjct: 437 SVSLDEVQTALNSKELNERKEKKSSASGEGLTARGKTFKKDSEFDKKKQKPENQKNGEGN 258
Query: 169 PAEIVCYKCGEKGHKSNVCNGEEK 192
+I CY C ++GH VC +K
Sbjct: 257 IFKIRCYHCKKEGHTRKVCPERQK 186
>TC217111 similar to UP|P93188 (P93188) NADPH-dependent HC-toxin reductase ,
partial (23%)
Length = 1312
Score = 29.6 bits (65), Expect = 1.4
Identities = 17/62 (27%), Positives = 31/62 (49%)
Frame = +1
Query: 64 AAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYEED 123
A+ ++ L + HY+ EF+ ++E+GL+ DIK A ++L +Y+ D
Sbjct: 1027 ASSYISLEEMANHYALHYPEFNVKQEYEDGLKKDIKWA--------STKLCDKGFVYKYD 1182
Query: 124 TK 125
K
Sbjct: 1183 AK 1188
>TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein-1, partial
(77%)
Length = 883
Score = 28.5 bits (62), Expect = 3.2
Identities = 9/16 (56%), Positives = 11/16 (68%)
Frame = +3
Query: 174 CYKCGEKGHKSNVCNG 189
CY CGE GH + C+G
Sbjct: 435 CYNCGESGHLARDCSG 482
Score = 26.9 bits (58), Expect = 9.4
Identities = 9/18 (50%), Positives = 10/18 (55%)
Frame = +3
Query: 210 GDIVCFNCDEEGHLTSQC 227
G C+NC E GHL C
Sbjct: 423 GGGACYNCGESGHLARDC 476
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,408,409
Number of Sequences: 63676
Number of extensions: 185962
Number of successful extensions: 1151
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 1017
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1103
length of query: 264
length of database: 12,639,632
effective HSP length: 96
effective length of query: 168
effective length of database: 6,526,736
effective search space: 1096491648
effective search space used: 1096491648
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC137553.2


