

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
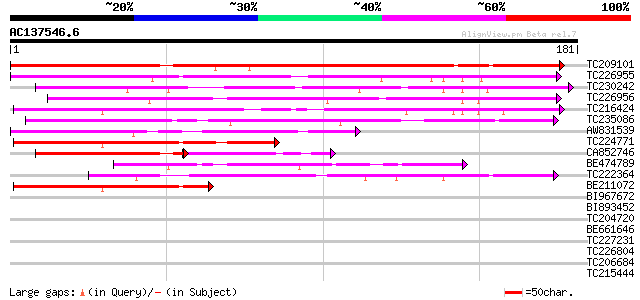
Query= AC137546.6 - phase: 0
(181 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC209101 similar to UP|Q9FQZ6 (Q9FQZ6) Avr9/Cf-9 rapidly elicite... 153 5e-38
TC226955 similar to UP|Q9FQZ6 (Q9FQZ6) Avr9/Cf-9 rapidly elicite... 118 2e-27
TC230242 weakly similar to UP|Q9FQZ6 (Q9FQZ6) Avr9/Cf-9 rapidly ... 112 7e-26
TC226956 similar to UP|Q9FQZ6 (Q9FQZ6) Avr9/Cf-9 rapidly elicite... 112 1e-25
TC216424 similar to GB|AAD12688.1|4220461|F5F19 ESTs gb|T75642 a... 88 3e-18
TC235086 75 2e-14
AW831539 68 3e-12
TC224771 64 4e-11
CA852746 weakly similar to GP|19528011|gb| AT20635p {Drosophila ... 61 5e-11
BE474789 similar to GP|9279805|dbj AmiB {Dictyostelium discoideu... 62 1e-10
TC222364 similar to GB|AAD12688.1|4220461|F5F19 ESTs gb|T75642 a... 60 6e-10
BE211072 58 2e-09
BI967672 35 0.019
BI893452 33 0.056
TC204720 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17... 32 0.21
BE661646 weakly similar to PIR|AG0096|AG00 phosphoenolpyruvate-p... 31 0.28
TC227231 similar to UP|Q6ZI79 (Q6ZI79) Proline-rich family prote... 31 0.28
TC226804 homologue to UP|Q6YH15 (Q6YH15) 5-enolpyruvylshikimate-... 28 2.4
TC206684 weakly similar to UP|Q9FYL8 (Q9FYL8) F21J9.11 (At1g2445... 27 4.0
TC215444 homologue to UP|Q39013 (Q39013) ATAF1 protein, partial ... 27 5.3
>TC209101 similar to UP|Q9FQZ6 (Q9FQZ6) Avr9/Cf-9 rapidly elicited protein
146, partial (29%)
Length = 1428
Score = 153 bits (386), Expect = 5e-38
Identities = 91/181 (50%), Positives = 119/181 (65%), Gaps = 4/181 (2%)
Frame = +3
Query: 1 MEIEARQGVVAKKLWNMVRVLLFIIRKGIAKSKTMVHLNLILKRGKLAGKALINTLMLNH 60
MEIEA + VVAKK+WNMVRVL F++RKGIAKSK MV +L+LKRGKLA +N L+LNH
Sbjct: 33 MEIEASRPVVAKKVWNMVRVLFFMLRKGIAKSKIMVEFHLMLKRGKLA----LNNLILNH 200
Query: 61 QLYH---SSFTCRSDNNS-FISPCEYEFSCRNTPANPLRHSSRRFSKSRRQRHNDFSIMN 116
Y+ S FTCRS ++S FISP +YEFSC N+PA P + ++R +KS I+N
Sbjct: 201 HYYNMQTSFFTCRSHHHSTFISPSDYEFSCSNSPAIPTKRTNRFSTKSLSSVQKVLEILN 380
Query: 117 NNIAVQKVFIEMMLNNEKVEAVANSPLAPFTLEDEGNCHQVDIAAEEFINNFYKELNRQN 176
N + F + V + + +PF L+DE + QVD+AAEEFI NFYK+LN Q
Sbjct: 381 NENNINSPFPGFGKSPASVRQLRVTD-SPFPLKDEED-SQVDVAAEEFIKNFYKDLNLQK 554
Query: 177 R 177
+
Sbjct: 555 K 557
>TC226955 similar to UP|Q9FQZ6 (Q9FQZ6) Avr9/Cf-9 rapidly elicited protein
146, partial (20%)
Length = 934
Score = 118 bits (295), Expect = 2e-27
Identities = 81/206 (39%), Positives = 116/206 (55%), Gaps = 30/206 (14%)
Frame = +1
Query: 1 MEIEARQGVVAKKLWNMVRVLLFIIRKGIAKSKTMVHLNLILKR-GKLAGKALINTLMLN 59
+E+E+ +AKK+W+MVRV+LF++RKGI K K M+ LN++LKR GKLAGKA+ N LM +
Sbjct: 88 IEMESNASALAKKVWSMVRVVLFMLRKGITKGKLMMDLNMMLKRRGKLAGKAIAN-LMSS 264
Query: 60 HQLYHSSFTCRSDNNSFISPCEYEFSCRNTPANPLRHSSRRFSKSRRQRHNDFSIMNN-- 117
H ++ + S + F +P EYEFSC NTP H+ + + RH+ F+ ++
Sbjct: 265 HHHHNRGGSHNSHHLKFSAPREYEFSCSNTP-----HNFFFPTGGKHHRHHFFACVHAPP 429
Query: 118 -------NIAVQKVFIEMMLNNE----KVEA------VANSPL--------APFTLE--D 150
+ K +EM+ NN KVEA SP+ +PF L D
Sbjct: 430 TQDDDVVTVNAMKAVMEMLNNNNNDAVKVEASPALPGFGRSPMVRQLRVTDSPFPLRDAD 609
Query: 151 EGNCHQVDIAAEEFINNFYKELNRQN 176
+ N H VD AAE+FI FYKEL +Q+
Sbjct: 610 QDNDHLVDKAAEDFIKRFYKELRKQD 687
>TC230242 weakly similar to UP|Q9FQZ6 (Q9FQZ6) Avr9/Cf-9 rapidly elicited
protein 146, partial (30%)
Length = 713
Score = 112 bits (281), Expect = 7e-26
Identities = 81/197 (41%), Positives = 111/197 (56%), Gaps = 25/197 (12%)
Frame = +2
Query: 9 VVAKKLWNMVRVLLFIIRKGIAKSKTMV--HLNLILKRGKLAG-KALINTLMLNHQLYHS 65
VVA+KLWN+VR++ ++RKGI+KSK +LNL KRGKLA KA+ NT++
Sbjct: 92 VVARKLWNIVRIVFLMLRKGISKSKLAADFNLNLSFKRGKLAATKAIANTIL-------- 247
Query: 66 SFTCRSDNNSFISPCEYEFSCRNTPANPLRHSSRRFSKSRRQRHN------DFSIMNNNI 119
RS N+ + P +YEFSC N+PA L H +R + R H+ F ++
Sbjct: 248 ---HRSQNDDAVYPRDYEFSCSNSPA--LFHVIKRSTNKHRHHHHRCTTTTKFPHHDDVS 412
Query: 120 AVQKVFIEMMLNNEKVEA-----VANSPL---------APFTLEDE--GNCHQVDIAAEE 163
VQKV +LNN KVEA SP+ +PF L+DE G+ +QVD+AA+E
Sbjct: 413 TVQKVL--EILNNNKVEASPLPGFGKSPIGRKLIRITDSPFPLKDEEGGDENQVDVAADE 586
Query: 164 FINNFYKELNRQNRTIA 180
FI FYK+LN Q + A
Sbjct: 587 FIKRFYKDLNLQQKMAA 637
>TC226956 similar to UP|Q9FQZ6 (Q9FQZ6) Avr9/Cf-9 rapidly elicited protein
146, partial (33%)
Length = 835
Score = 112 bits (280), Expect = 1e-25
Identities = 76/187 (40%), Positives = 101/187 (53%), Gaps = 23/187 (12%)
Frame = +2
Query: 13 KLWNMVRVLLFIIRKGIAKSKTMVHLNLILK-RGKLAGKALINTLMLNHQLYHSSFTCRS 71
K+W+MVRVLLF++RKGI+K K M+ LN++LK RGKLAGKA+ N + NH S
Sbjct: 47 KVWSMVRVLLFMLRKGISKGKLMMDLNMMLKRRGKLAGKAIANLMSHNHNHRGS----HK 214
Query: 72 DNNSFISPCEYEFSCRNTPANPLRHSSRR----FSKSRRQRHNDFSIMNNNIAVQKVFIE 127
F +P EYEFSC NTP N + +R F+ D ++ N K +E
Sbjct: 215 SQLRFSAPREYEFSCSNTPHNLFPTAGKRHRHFFACVHAPPTQDDDVVTVN--AMKAVLE 388
Query: 128 MMLNNEKVEAVANSPL----------------APFTL--EDEGNCHQVDIAAEEFINNFY 169
M+ NN+ V+ A+ L +PF L D N HQVD AAEEFI FY
Sbjct: 389 MLNNNDVVKVEASPALPGFGRSPIVRQLRVTDSPFPLRDSDHDNDHQVDKAAEEFIKRFY 568
Query: 170 KELNRQN 176
K+L +Q+
Sbjct: 569 KDLKKQD 589
>TC216424 similar to GB|AAD12688.1|4220461|F5F19 ESTs gb|T75642 and
gb|AA650997 come from this gene. {Arabidopsis thaliana;}
, partial (15%)
Length = 1166
Score = 87.8 bits (216), Expect = 3e-18
Identities = 70/216 (32%), Positives = 108/216 (49%), Gaps = 40/216 (18%)
Frame = +3
Query: 2 EIEARQGVVAKKLWNMVRVLLFIIRKG-IAKSKTMVHLNLILKRGKLAGKALINTLMLNH 60
++EA V+AKKLWN++R+ F+IRKG ++K K ++ +NL++K+GKL K++ N + +H
Sbjct: 267 KMEAHSPVIAKKLWNVLRITFFMIRKGLVSKRKLIMDMNLMMKKGKLLRKSMSNFMSSHH 446
Query: 61 QLYHSSFTCRSDNNSFISPCEYEFSCRNTPANPLRHSSRRFSKSRRQRHNDFSIMNNNIA 120
++S R EYEFSC N+P NP+ F +R+ H +F MN
Sbjct: 447 HHHYSKSLARGYGMQ-----EYEFSCSNSP-NPVF-----FHVPKRKHHFNFPCMNTPEV 593
Query: 121 VQKVF---IEMMLNNEKVEAVAN-----------------SPL----APFTL-------E 149
V++ + ++ NN E N SPL +PF++ E
Sbjct: 594 VEEPNNRPVVLVPNNMTPEYTFNLRFDNINASDFAPGERKSPLLSSSSPFSVRISNYSSE 773
Query: 150 DEGNCHQ--------VDIAAEEFINNFYKELNRQNR 177
DE N Q VD AE+FI FY++L Q+R
Sbjct: 774 DEENEEQEGGNGNGHVDDQAEDFIRRFYEQLRMQSR 881
>TC235086
Length = 696
Score = 75.1 bits (183), Expect = 2e-14
Identities = 56/177 (31%), Positives = 86/177 (47%), Gaps = 7/177 (3%)
Frame = +1
Query: 6 RQGVVAKKLWNMVRVLLFIIRKGIAKSKTMVHLNLILKRGKLAGKALINTLMLNHQLYHS 65
R +VA+K++ VR + F ++ G++KSK + ++KRGK GKAL N ++ H H
Sbjct: 52 RPPLVAEKVFKTVRFIGFFLQNGVSKSKVFHDVQDVMKRGKNIGKAL-NDIVARH---HQ 219
Query: 66 SFTC--RSDNNSFISPCEYEFSCRNTPANPLRHSSRRFSKS-----RRQRHNDFSIMNNN 118
+ TC R + +F+SP EY+FSC +P R+ S + R H +
Sbjct: 220 ALTCRPRDAHVAFVSPLEYQFSCSGSPPRTSHAGRRKLSPASAAVRRSPGHRAVRMCRGG 399
Query: 119 IAVQKVFIEMMLNNEKVEAVANSPLAPFTLEDEGNCHQVDIAAEEFINNFYKELNRQ 175
A E+ + +S + F E + H VD AAEEFI FY++L Q
Sbjct: 400 GAADGTI-------ERRVKITDSVASMFNDTAEKDFH-VDEAAEEFIARFYRDLRLQ 546
>AW831539
Length = 397
Score = 67.8 bits (164), Expect = 3e-12
Identities = 43/113 (38%), Positives = 66/113 (58%), Gaps = 1/113 (0%)
Frame = +2
Query: 1 MEIEARQGVVAKKLWNMVRVLLFIIRKGIAKSKTMVHL-NLILKRGKLAGKALINTLMLN 59
+E+E+ V+AKK +++ V LF++RKGI+K K M+HL N++LKR GKA+ N
Sbjct: 14 IEMESNTPVIAKKELSIIHVALFMLRKGISKGKLMMHLNNMMLKR---PGKAIANL---- 172
Query: 60 HQLYHSSFTCRSDNNSFISPCEYEFSCRNTPANPLRHSSRRFSKSRRQRHNDF 112
++H + ++ F + EYEFSC NTP N + S ++R RH F
Sbjct: 173 --MFHHNHGVSGNSLHFSAAREYEFSCSNTPNN---FFAAIASSNKRHRHIPF 316
>TC224771
Length = 475
Score = 63.9 bits (154), Expect = 4e-11
Identities = 37/86 (43%), Positives = 56/86 (65%), Gaps = 1/86 (1%)
Frame = +1
Query: 2 EIEARQGVVAKKLWNMVRVLLFIIRKG-IAKSKTMVHLNLILKRGKLAGKALINTLMLNH 60
E+E VVAK+LWN++RV F+IRKG I+K K ++ +NL++K+GK+ K+L N LM +H
Sbjct: 229 EMEVHSPVVAKRLWNVLRVTFFMIRKGLISKRKMIMDMNLMMKKGKVVRKSLSN-LMSSH 405
Query: 61 QLYHSSFTCRSDNNSFISPCEYEFSC 86
+H+ +S + YEFSC
Sbjct: 406 HHHHNH---KSVTHGGGLGVHYEFSC 474
>CA852746 weakly similar to GP|19528011|gb| AT20635p {Drosophila
melanogaster}, partial (7%)
Length = 516
Score = 61.2 bits (147), Expect(2) = 5e-11
Identities = 32/49 (65%), Positives = 39/49 (79%)
Frame = +3
Query: 9 VVAKKLWNMVRVLLFIIRKGIAKSKTMVHLNLILKRGKLAGKALINTLM 57
VVAKK+WNMVR+L F++RKGIAKSK M +L+LKRGKLA +N LM
Sbjct: 75 VVAKKVWNMVRMLFFMLRKGIAKSKIMA*FHLMLKRGKLA----LNNLM 209
Score = 22.3 bits (46), Expect(2) = 5e-11
Identities = 16/49 (32%), Positives = 24/49 (48%)
Frame = +1
Query: 56 LMLNHQLYHSSFTCRSDNNSFISPCEYEFSCRNTPANPLRHSSRRFSKS 104
+ LNH Y +SF+C + + + FS +T P RRFS+S
Sbjct: 211 MQLNHHHYITSFSCSNSHTKRTNHFTTRFS--STKY*P---QIRRFSRS 342
>BE474789 similar to GP|9279805|dbj AmiB {Dictyostelium discoideum}, partial
(0%)
Length = 368
Score = 62.0 bits (149), Expect = 1e-10
Identities = 45/116 (38%), Positives = 69/116 (58%), Gaps = 3/116 (2%)
Frame = +1
Query: 34 TMVHLNLILKRGKLAG-KALINTLMLNHQLYHSSFTCRSDNNSFISPCEYEFSCRNTPA- 91
T +LNL+LKRGKLA KA+ NT++ H L+ S ++D+++ P +YEFSC N+PA
Sbjct: 1 TSFNLNLLLKRGKLAATKAIANTILTLH-LHRS----QNDDDAVYYPRDYEFSCSNSPAL 165
Query: 92 -NPLRHSSRRFSKSRRQRHNDFSIMNNNIAVQKVFIEMMLNNEKVEAVANSPLAPF 146
+ ++ S+ + + RR RH D + VQKV +E++ NN + SPL F
Sbjct: 166 FHVIKRSTNKHHR-RRHRHYDDDVS----TVQKV-LEILNNNNNNKVATASPLPGF 315
>TC222364 similar to GB|AAD12688.1|4220461|F5F19 ESTs gb|T75642 and
gb|AA650997 come from this gene. {Arabidopsis thaliana;}
, partial (19%)
Length = 719
Score = 60.1 bits (144), Expect = 6e-10
Identities = 56/180 (31%), Positives = 82/180 (45%), Gaps = 30/180 (16%)
Frame = +1
Query: 26 RKGIAKSKTMVHLN-LILKRGKLAGKALINTLMLNHQLYHSSFTCRSDNNSFISPCEYEF 84
RKGI K K M+ L ++LKR + + ++H + ++ F + EYEF
Sbjct: 1 RKGIXKGKLMMXLXXMMLKRXXXX---------IANLMFHHNHGVSGNSLHFSAAREYEF 153
Query: 85 SCRNTPANPLRHSSRRFSKSRRQRHNDF-------SIMNNNIAVQ---KVFIEMMLNNEK 134
SC NTP N + S ++R RH F +++++A K +EM+ N
Sbjct: 154 SCSNTPNNFFXAIA---SSNKRHRHIPFFTCAHAPPTLDDDVATVNAVKGVLEMLNNETV 324
Query: 135 VEA-------------------VANSPLAPFTLEDEGNCHQVDIAAEEFINNFYKELNRQ 175
VEA V +SP ED+ + +QVD AAEEFI FYKEL +Q
Sbjct: 325 VEASPAALPGFGRSPMVRQLIRVTDSPFPLVESEDDKD-NQVDKAAEEFIKRFYKELRKQ 501
>BE211072
Length = 408
Score = 58.2 bits (139), Expect = 2e-09
Identities = 30/65 (46%), Positives = 47/65 (72%), Gaps = 1/65 (1%)
Frame = +3
Query: 2 EIEARQGVVAKKLWNMVRVLLFIIRKG-IAKSKTMVHLNLILKRGKLAGKALINTLMLNH 60
E+E V AK+LWN++RV F+IRKG I+K K ++ +NL++K+GK+ K+L N LM +H
Sbjct: 189 EMEVHSPVGAKRLWNVLRVTFFMIRKGLISKRKMIMDMNLMMKKGKVVRKSLSN-LMSSH 365
Query: 61 QLYHS 65
+H+
Sbjct: 366 HHHHN 380
>BI967672
Length = 335
Score = 35.0 bits (79), Expect = 0.019
Identities = 15/26 (57%), Positives = 20/26 (76%)
Frame = -3
Query: 155 HQVDIAAEEFINNFYKELNRQNRTIA 180
+QVD+AAEEFI FYK+L+ Q + A
Sbjct: 330 NQVDVAAEEFIKRFYKDLDLQQKMAA 253
>BI893452
Length = 425
Score = 33.5 bits (75), Expect = 0.056
Identities = 21/78 (26%), Positives = 36/78 (45%)
Frame = -2
Query: 24 IIRKGIAKSKTMVHLNLILKRGKLAGKALINTLMLNHQLYHSSFTCRSDNNSFISPCEYE 83
+I + +T++H +L+ G LA K+ + ++ H Y S + N+ F+ PCEY+
Sbjct: 319 VITNNLYFPRTIIHERNLLQEGNLAKKSDMKRVLY*HHQY-SFAEFKVSNSFFLKPCEYK 143
Query: 84 FSCRNTPANPLRHSSRRF 101
R S RF
Sbjct: 142 MPSSIC*IIQYRRSEGRF 89
>TC204720 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17H15.1
{Arabidopsis thaliana;} , partial (31%)
Length = 1936
Score = 31.6 bits (70), Expect = 0.21
Identities = 27/91 (29%), Positives = 40/91 (43%), Gaps = 15/91 (16%)
Frame = +1
Query: 23 FIIRKGIAKSKTMVHLNLILKRGK-------LAGKALI---NTLML-----NHQLYHSSF 67
++I+ +A+ +H L++ K LAG L+ N L++ H L H S
Sbjct: 595 WLIKSSVARIVREIHQCLVVILSKVTKLGHPLAGVPLLLLCNNLVMAMCSQEHTLVHPSI 774
Query: 68 TCRSDNNSFISPCEYEFSCRNTPANPLRHSS 98
TC S+N I P T +PL HSS
Sbjct: 775 TCLSNNMQAILPSHLVDILPPTGISPLPHSS 867
>BE661646 weakly similar to PIR|AG0096|AG00 phosphoenolpyruvate-protein
phosphotransferase (EC 2.7.3.9) [imported] - Yersinia
pestis, partial (2%)
Length = 813
Score = 31.2 bits (69), Expect = 0.28
Identities = 22/68 (32%), Positives = 32/68 (46%), Gaps = 10/68 (14%)
Frame = +1
Query: 60 HQLYHSSFTCRSDNNSFISPCEYEF----SC--RNTPANPLRHSSRRFSKSRR----QRH 109
+ +Y++SFTCR +N PC E +C R A S RR S S R R+
Sbjct: 13 YPIYNNSFTCRKENCGL*YPC*EEMYSSTTCRGRKPAATAAASSGRRTSNSTRLEFENRY 192
Query: 110 NDFSIMNN 117
+D ++ N
Sbjct: 193 SDKRVIRN 216
>TC227231 similar to UP|Q6ZI79 (Q6ZI79) Proline-rich family protein-like,
partial (45%)
Length = 1060
Score = 31.2 bits (69), Expect = 0.28
Identities = 28/93 (30%), Positives = 40/93 (42%), Gaps = 5/93 (5%)
Frame = +3
Query: 55 TLMLNHQLYHSSFTCRSDNNSFISPCEYEFSCRNTPANPLRHSSRRFSKSRRQRHNDFSI 114
TL+L L+HSS + ++ Y FS R ++S RRFS + +D S
Sbjct: 135 TLILEFFLFHSSLVMACALQATLAANTYAFSSRRFSLKHQKNSKRRFSLFTVRADSDDSD 314
Query: 115 MNN-----NIAVQKVFIEMMLNNEKVEAVANSP 142
N + V KV +E L EK + V P
Sbjct: 315 CNEEECAPDKEVGKVSVE-WLAGEKTKVVGTFP 410
>TC226804 homologue to UP|Q6YH15 (Q6YH15) 5-enolpyruvylshikimate-3-phosphate
synthase (Fragment), partial (43%)
Length = 630
Score = 28.1 bits (61), Expect = 2.4
Identities = 14/45 (31%), Positives = 22/45 (48%)
Frame = -1
Query: 59 NHQLYHSSFTCRSDNNSFISPCEYEFSCRNTPANPLRHSSRRFSK 103
NH + + F C S + F + C + S N+P + +SS SK
Sbjct: 264 NHTVIRTFFNCCS*LSEFCADCYHPLSLFNSPTCHISYSSGTISK 130
>TC206684 weakly similar to UP|Q9FYL8 (Q9FYL8) F21J9.11
(At1g24450/F21J9_210), partial (57%)
Length = 798
Score = 27.3 bits (59), Expect = 4.0
Identities = 24/80 (30%), Positives = 29/80 (36%), Gaps = 25/80 (31%)
Frame = +3
Query: 60 HQLYHSSFTCRSDNNSFI--SPCEYEFSCRNTPANP-----------------------L 94
H L H R+ NNS + SP FS N P NP L
Sbjct: 99 HPLSHCDPPPRTRNNSNLQTSPQTSLFSLFNRPRNPPETTRVHLQEHRSSSSRHDTRFLL 278
Query: 95 RHSSRRFSKSRRQRHNDFSI 114
R + F SRRQRH + +
Sbjct: 279 RRKQQSFRHSRRQRHRNLRL 338
>TC215444 homologue to UP|Q39013 (Q39013) ATAF1 protein, partial (59%)
Length = 1198
Score = 26.9 bits (58), Expect = 5.3
Identities = 18/53 (33%), Positives = 27/53 (49%)
Frame = -3
Query: 53 INTLMLNHQLYHSSFTCRSDNNSFISPCEYEFSCRNTPANPLRHSSRRFSKSR 105
INT ++ + H+S + +S S C + F C N P R SR F++SR
Sbjct: 878 INTSLI*YT--HNSSCSYKNLSSSSSWCFFVFFCNNRLHEPARTHSRIFTQSR 726
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,917,781
Number of Sequences: 63676
Number of extensions: 105251
Number of successful extensions: 672
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 654
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 656
length of query: 181
length of database: 12,639,632
effective HSP length: 91
effective length of query: 90
effective length of database: 6,845,116
effective search space: 616060440
effective search space used: 616060440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC137546.6


