

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137546.2 - phase: 0
(623 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
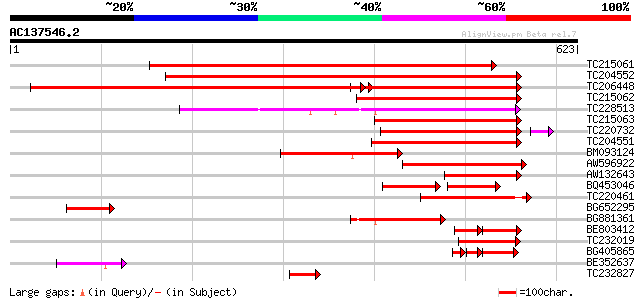
Score E
Sequences producing significant alignments: (bits) Value
TC215061 similar to UP|O24550 (O24550) Malate dehydrogenase , p... 679 0.0
TC204552 homologue to UP|Q9M4Q9 (Q9M4Q9) NADP-dependent malic pr... 660 0.0
TC206448 homologue to GB|CAA56354.1|510876|PVME1G NADP dependent... 644 0.0
TC215062 similar to UP|MAOX_VITVI (P51615) NADP-dependent malic ... 323 1e-88
TC228513 similar to UP|MAOM_SOLTU (P37221) NAD-dependent malic e... 298 5e-81
TC215063 similar to UP|MAOX_VITVI (P51615) NADP-dependent malic ... 280 1e-75
TC220732 similar to UP|Q6PMI1 (Q6PMI1) NADP-dependent malic enzy... 273 1e-73
TC204551 homologue to UP|Q9M4Q9 (Q9M4Q9) NADP-dependent malic pr... 263 1e-70
BM093124 231 7e-61
AW596922 similar to GP|8118507|gb| NADP-dependent malic protein ... 147 1e-35
AW132643 similar to GP|8118507|gb| NADP-dependent malic protein ... 139 4e-33
BQ453046 similar to GP|8894554|emb NAD-dependent malic enzyme (m... 65 2e-22
TC220461 similar to UP|Q9LEN2 (Q9LEN2) NAD-dependent malic enzym... 97 3e-20
BG652295 similar to SP|P51615|MAOX_ NADP-dependent malic enzyme ... 82 5e-16
BG881361 homologue to GP|8894554|emb NAD-dependent malic enzyme ... 77 2e-14
BE803412 69 6e-12
TC232019 homologue to UP|Q9LEN2 (Q9LEN2) NAD-dependent malic enz... 64 2e-10
BG405865 homologue to GP|8118507|gb|A NADP-dependent malic prote... 63 4e-10
BE352637 similar to SP|P37225|MAON NAD-dependent malic enzyme 59... 52 1e-06
TC232827 similar to UP|Q94G60 (Q94G60) Sucrose synthase , parti... 51 1e-06
>TC215061 similar to UP|O24550 (O24550) Malate dehydrogenase , partial (58%)
Length = 1146
Score = 679 bits (1751), Expect = 0.0
Identities = 335/381 (87%), Positives = 361/381 (93%)
Frame = +1
Query: 154 YKRPQGLYISLKEKGKILEVLKNWPERSIQVIVVTDGERILGLGDLGCQGMGIPVGKLAL 213
+KRPQGL+ISLKEKGK+LEVLKNWPE+SIQVIVVTDGERILGLGDLGCQGMGIPVGKLAL
Sbjct: 1 FKRPQGLFISLKEKGKVLEVLKNWPEKSIQVIVVTDGERILGLGDLGCQGMGIPVGKLAL 180
Query: 214 YSALGGVRPSACLPVTIDVGTNNEKLLNDEFYIGLRQKRATGKEYYDLLHEFMTAVKQNY 273
Y+ALGGVRPSACLP+TIDVGTNNEKLLNDEFYIGLRQKRATG+EY +LL EFMTAVKQNY
Sbjct: 181 YTALGGVRPSACLPITIDVGTNNEKLLNDEFYIGLRQKRATGQEYSELLQEFMTAVKQNY 360
Query: 274 GEKVLVQFEDFANHNAFELLAKYGTTHLVFNDDIQGTAAVVLAGVVASLKLIGGTLPEHT 333
GEKVL+QFEDFANHNAFELLAKYGTTHLVFNDDIQGTA+VVLAGVVA+LKLIGG L +HT
Sbjct: 361 GEKVLIQFEDFANHNAFELLAKYGTTHLVFNDDIQGTASVVLAGVVAALKLIGGNLADHT 540
Query: 334 FLFLGAGEAGTGIAELIALEMSKQTKAPIEESRKKIWLVDSKGLIVSSRANSLQHFKKPW 393
FLFLGAGEAGTGIAELIALEMSKQTK PIEE+RKKIWLVDSKGLIV SR SLQHFK+PW
Sbjct: 541 FLFLGAGEAGTGIAELIALEMSKQTKTPIEETRKKIWLVDSKGLIVGSRKASLQHFKQPW 720
Query: 394 AHEHEPVSTLLDAVKIIKPTVLIGSSGVGKTFTKEVVEAMTEINKIPLILALSNPTSQSE 453
AHEHEPV +LL+AVK+IKPTVLIGSSGVGKTFTKEV+EA+T IN+ PL+LALSNPTSQSE
Sbjct: 721 AHEHEPVGSLLEAVKVIKPTVLIGSSGVGKTFTKEVIEAVTSINEKPLVLALSNPTSQSE 900
Query: 454 CTAEEAYTWSEGRAIFASGSPFDPVEYKGKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHD 513
CTAEEAY WSEGRAIFASGSPFDPVEYKGK YYSGQ+NNAYIFPGFGLG+V+SGAIRVHD
Sbjct: 901 CTAEEAYQWSEGRAIFASGSPFDPVEYKGKVYYSGQANNAYIFPGFGLGLVISGAIRVHD 1080
Query: 514 DMLLAASEALAKQVTEENYKK 534
DML AS ALAK V E Y++
Sbjct: 1081DMLSTASVALAKLVYNEYYEQ 1143
>TC204552 homologue to UP|Q9M4Q9 (Q9M4Q9) NADP-dependent malic protein ,
partial (66%)
Length = 1640
Score = 660 bits (1703), Expect = 0.0
Identities = 322/391 (82%), Positives = 361/391 (91%)
Frame = +3
Query: 172 EVLKNWPERSIQVIVVTDGERILGLGDLGCQGMGIPVGKLALYSALGGVRPSACLPVTID 231
EVL+NWPE++IQVIVVTDGERILGLGDLGCQGMGIPVGKL+LY+ALGGVRPSACLP+TID
Sbjct: 3 EVLRNWPEKNIQVIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSACLPITID 182
Query: 232 VGTNNEKLLNDEFYIGLRQKRATGKEYYDLLHEFMTAVKQNYGEKVLVQFEDFANHNAFE 291
VGTNNEKLLNDE YIGL+Q+RATG+EY +L+HEFMTAVKQ YGEKVL+QFEDFANHNAF+
Sbjct: 183 VGTNNEKLLNDELYIGLKQRRATGQEYAELMHEFMTAVKQTYGEKVLIQFEDFANHNAFD 362
Query: 292 LLAKYGTTHLVFNDDIQGTAAVVLAGVVASLKLIGGTLPEHTFLFLGAGEAGTGIAELIA 351
LL KY +THLVFNDDIQGTA+VVLAG+VASLKL+GG L +H FLFLGAGEAGTGIAELIA
Sbjct: 363 LLEKYRSTHLVFNDDIQGTASVVLAGLVASLKLVGGNLADHRFLFLGAGEAGTGIAELIA 542
Query: 352 LEMSKQTKAPIEESRKKIWLVDSKGLIVSSRANSLQHFKKPWAHEHEPVSTLLDAVKIIK 411
LE SKQT AP+EE RK IWLVDSKGLIVSSR +SLQHFKKPWAHEHEPV LLDAV IK
Sbjct: 543 LETSKQTNAPLEEVRKNIWLVDSKGLIVSSRKDSLQHFKKPWAHEHEPVKNLLDAVNKIK 722
Query: 412 PTVLIGSSGVGKTFTKEVVEAMTEINKIPLILALSNPTSQSECTAEEAYTWSEGRAIFAS 471
PTVLIG+SG G+TFTKEV+EAM IN+ P+IL+LSNPTSQSECTAEEAY WS+GRAIFAS
Sbjct: 723 PTVLIGTSGQGRTFTKEVIEAMASINERPIILSLSNPTSQSECTAEEAYKWSQGRAIFAS 902
Query: 472 GSPFDPVEYKGKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHDDMLLAASEALAKQVTEEN 531
GSPF PVEY+GK + GQ+NNAYIFPGFGLG++MSG IRVHDD+LLAASEALA QV++EN
Sbjct: 903 GSPFPPVEYEGKVFVPGQANNAYIFPGFGLGLIMSGTIRVHDDLLLAASEALAAQVSQEN 1082
Query: 532 YKKGLTYPPFSDIRKISANIAANVAAKAYEL 562
+ KGL YPPF++IRKISA+IAANVAAKAYEL
Sbjct: 1083FDKGLIYPPFTNIRKISAHIAANVAAKAYEL 1175
>TC206448 homologue to GB|CAA56354.1|510876|PVME1G NADP dependent malic
enzyme {Phaseolus vulgaris;} , partial (97%)
Length = 2208
Score = 644 bits (1662), Expect(2) = 0.0
Identities = 311/368 (84%), Positives = 348/368 (94%)
Frame = +3
Query: 24 GGGVEDVYGEDCATEDQLVTPWTFSVASGYSLLRDPQYNKGLAFTEKERDAHYLRGLLPP 83
GGGV D+YGEDCATEDQL+TPWTFSVASG SLLRDP+YNKGLAFTE ERDAHYLRGLLPP
Sbjct: 159 GGGVRDLYGEDCATEDQLITPWTFSVASGCSLLRDPRYNKGLAFTEGERDAHYLRGLLPP 338
Query: 84 TVSSQQLQEKKLMHNIRQYEVPLQKYVAMMDLQERNERLFYKLLIDNVEELLPIVYTPVV 143
+ +Q+LQEK+LMHN+RQYEVPL +Y+AMMDLQERNERLFYKLLI+NVEELLP+VYTP V
Sbjct: 339 AIFNQELQEKRLMHNLRQYEVPLHRYMAMMDLQERNERLFYKLLINNVEELLPVVYTPTV 518
Query: 144 GEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPERSIQVIVVTDGERILGLGDLGCQG 203
GEACQKYGSI++RPQGLYISLKEKGKILEVLKNWPE+SIQVIVVTDGERILGLGDLGCQG
Sbjct: 519 GEACQKYGSIFRRPQGLYISLKEKGKILEVLKNWPEKSIQVIVVTDGERILGLGDLGCQG 698
Query: 204 MGIPVGKLALYSALGGVRPSACLPVTIDVGTNNEKLLNDEFYIGLRQKRATGKEYYDLLH 263
MGIPVGKL+LY+ALGGVRPS+CLP+TIDVGTNNEKLLNDEFYIGL+QKRATG+EY +LL
Sbjct: 699 MGIPVGKLSLYTALGGVRPSSCLPITIDVGTNNEKLLNDEFYIGLKQKRATGQEYAELLE 878
Query: 264 EFMTAVKQNYGEKVLVQFEDFANHNAFELLAKYGTTHLVFNDDIQGTAAVVLAGVVASLK 323
EFM AVKQNYGEKVL+QFEDFANHNAF+LL KY ++HLVFNDDIQGTA+VVLAG++ASLK
Sbjct: 879 EFMHAVKQNYGEKVLIQFEDFANHNAFDLLEKYSSSHLVFNDDIQGTASVVLAGLLASLK 1058
Query: 324 LIGGTLPEHTFLFLGAGEAGTGIAELIALEMSKQTKAPIEESRKKIWLVDSKGLIVSSRA 383
L+GGTL +HTFLFLGAGEAGTGIAELIALE+SKQTKAP+EE+RKKIWLVDSKG I SSR+
Sbjct: 1059LVGGTLVDHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKIWLVDSKGKIASSRS 1238
Query: 384 NSLQHFKK 391
+ +K
Sbjct: 1239RTTSPVQK 1262
Score = 310 bits (795), Expect = 9e-85
Identities = 153/188 (81%), Positives = 170/188 (90%)
Frame = +2
Query: 375 KGLIVSSRANSLQHFKKPWAHEHEPVSTLLDAVKIIKPTVLIGSSGVGKTFTKEVVEAMT 434
KGLIVSSR SLQ FKKPWAHEHEPV +LLDAVK IKPTVLIGSSG GKTFTKEVVE M
Sbjct: 1313 KGLIVSSRLESLQQFKKPWAHEHEPVKSLLDAVKAIKPTVLIGSSGAGKTFTKEVVETMA 1492
Query: 435 EINKIPLILALSNPTSQSECTAEEAYTWSEGRAIFASGSPFDPVEYKGKTYYSGQSNNAY 494
+N+ PLILALSNPTSQSECTAEEAYTWS+G+AIFASGSPFDPVEY+GK + GQ+NNAY
Sbjct: 1493 SLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVEYEGKLFVPGQANNAY 1672
Query: 495 IFPGFGLGIVMSGAIRVHDDMLLAASEALAKQVTEENYKKGLTYPPFSDIRKISANIAAN 554
IFPGFGLG+++SGAIRV D+MLLAASEALA QV++ENY KGL YPPFS+IRKISA+IAA
Sbjct: 1673 IFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFSNIRKISAHIAAK 1852
Query: 555 VAAKAYEL 562
VAAKAY+L
Sbjct: 1853 VAAKAYDL 1876
Score = 24.6 bits (52), Expect(2) = 0.0
Identities = 10/14 (71%), Positives = 10/14 (71%)
Frame = +1
Query: 386 LQHFKKPWAHEHEP 399
L FKK AHEHEP
Sbjct: 1246 LHQFKKT*AHEHEP 1287
>TC215062 similar to UP|MAOX_VITVI (P51615) NADP-dependent malic enzyme
(NADP-ME) , partial (36%)
Length = 971
Score = 323 bits (828), Expect = 1e-88
Identities = 158/181 (87%), Positives = 172/181 (94%)
Frame = +2
Query: 382 RANSLQHFKKPWAHEHEPVSTLLDAVKIIKPTVLIGSSGVGKTFTKEVVEAMTEINKIPL 441
R SLQHFK+PWAHEHEPV +LL+AVK+IKPTVLIGSSGVGKTFTKEV+EA+T IN+ PL
Sbjct: 2 RKASLQHFKQPWAHEHEPVGSLLEAVKVIKPTVLIGSSGVGKTFTKEVIEAVTSINEKPL 181
Query: 442 ILALSNPTSQSECTAEEAYTWSEGRAIFASGSPFDPVEYKGKTYYSGQSNNAYIFPGFGL 501
+LALSNPTSQSECTAEEAY WSEGRAIFASGSPFDPVEYKGK YYSGQ+NNAYIFPGFGL
Sbjct: 182 VLALSNPTSQSECTAEEAYEWSEGRAIFASGSPFDPVEYKGKVYYSGQANNAYIFPGFGL 361
Query: 502 GIVMSGAIRVHDDMLLAASEALAKQVTEENYKKGLTYPPFSDIRKISANIAANVAAKAYE 561
G+V+SGAIRVHDDMLLAASEALAK VTEENY+KGL YPPFS+IRKISANIAA+VAAKAYE
Sbjct: 362 GLVISGAIRVHDDMLLAASEALAKLVTEENYEKGLIYPPFSNIRKISANIAASVAAKAYE 541
Query: 562 L 562
L
Sbjct: 542 L 544
>TC228513 similar to UP|MAOM_SOLTU (P37221) NAD-dependent malic enzyme 62 kDa
isoform, mitochondrial precursor (NAD-ME) , partial
(69%)
Length = 1451
Score = 298 bits (763), Expect = 5e-81
Identities = 172/391 (43%), Positives = 238/391 (59%), Gaps = 16/391 (4%)
Frame = +2
Query: 187 VTDGERILGLGDLGCQGMGIPVGKLALYSALGGVRPSACLPVTIDVGTNNEKLLNDEFYI 246
VTDG RILGLGDLG QG+GI +GKL LY A G+ P LPV IDVGTNNEKLL D Y+
Sbjct: 2 VTDGSRILGLGDLGVQGIGIAIGKLDLYVAAAGINPQRVLPVMIDVGTNNEKLLEDPLYL 181
Query: 247 GLRQKRATGKEYYDLLHEFMTAVKQNYGEKVLVQFEDFANHNAFELLAKYGTTHLVFNDD 306
GL+Q R G +Y ++ EFM AV + V+VQFEDF + AF+LL +Y T+ +FNDD
Sbjct: 182 GLQQHRLDGDDYLAVVDEFMEAVFTRW-PNVIVQFEDFQSKWAFKLLQRYRNTYRMFNDD 358
Query: 307 IQGTAAVVLAGVVASLKLIGGTL---PEHTFLFLGAGEAGTGIAELIALEMSK---QTKA 360
+QGTA V +AG++ +++ G + P+ + GAG AG G+ M++ +
Sbjct: 359 VQGTAGVAIAGLLGAVRAQGRPMIDFPKQKIVVAGAGSAGIGVLNAARKTMARMLGNNEV 538
Query: 361 PIEESRKKIWLVDSKGLIVSSRANSLQHFKKPWAHEHEPV--------STLLDAVKIIKP 412
E ++ + W+VD++GLI R N + P+A + + ++L++ VK +KP
Sbjct: 539 AFESAKSQFWVVDAQGLITEGREN-IDPDALPFARNLKEMDRQGLREGASLVEVVKQVKP 715
Query: 413 TVLIGSSGVGKTFTKEVVEAMT-EINKIPLILALSNPTSQSECTAEEAYTWSEGRAIFAS 471
VL+G S VG F+KEV+EA+ + P I A+SNPT +ECTAEEA++ IFAS
Sbjct: 716 DVLLGLSAVGGLFSKEVLEALKGSTSTRPAIFAMSNPTKNAECTAEEAFSILGDNIIFAS 895
Query: 472 GSPFDPVEY-KGKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHDDMLLAASEALAKQVTEE 530
GSPF V+ G + Q NN Y+FPG GLG ++SGA V D ML AA+E LA ++EE
Sbjct: 896 GSPFSNVDLGNGHIGHCNQGNNMYLFPGIGLGTLLSGARIVSDGMLQAAAERLATYMSEE 1075
Query: 531 NYKKGLTYPPFSDIRKISANIAANVAAKAYE 561
KG+ +P S IR I+ +A V +A E
Sbjct: 1076EVLKGIIFPSTSRIRDITKQVATAVIKEAVE 1168
>TC215063 similar to UP|MAOX_VITVI (P51615) NADP-dependent malic enzyme
(NADP-ME) , partial (32%)
Length = 918
Score = 280 bits (717), Expect = 1e-75
Identities = 138/161 (85%), Positives = 153/161 (94%)
Frame = +2
Query: 402 TLLDAVKIIKPTVLIGSSGVGKTFTKEVVEAMTEINKIPLILALSNPTSQSECTAEEAYT 461
+LL+AVK+IKPTVLIGSSGVGKTFTKEV+EA+T IN+ PL+LALSNPTSQSECTAEEAY
Sbjct: 8 SLLEAVKVIKPTVLIGSSGVGKTFTKEVIEAVTSINEKPLVLALSNPTSQSECTAEEAYQ 187
Query: 462 WSEGRAIFASGSPFDPVEYKGKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHDDMLLAASE 521
WSEGRAIFASGSPFDPVEYKGK YYSGQ+NNAYIFPGFGLG+V+SGAIRVHDDMLLAASE
Sbjct: 188 WSEGRAIFASGSPFDPVEYKGKVYYSGQANNAYIFPGFGLGLVISGAIRVHDDMLLAASE 367
Query: 522 ALAKQVTEENYKKGLTYPPFSDIRKISANIAANVAAKAYEL 562
ALAK V+ ENY+KGL YPPFS+IR+ISANIAA+VA KAYEL
Sbjct: 368 ALAKLVSNENYEKGLIYPPFSNIREISANIAASVAGKAYEL 490
>TC220732 similar to UP|Q6PMI1 (Q6PMI1) NADP-dependent malic enzyme 3 ,
partial (32%)
Length = 662
Score = 273 bits (697), Expect(2) = 1e-73
Identities = 136/155 (87%), Positives = 146/155 (93%)
Frame = +1
Query: 408 KIIKPTVLIGSSGVGKTFTKEVVEAMTEINKIPLILALSNPTSQSECTAEEAYTWSEGRA 467
++IKPTVLIGSSGVG+TFTKEVVEAMT N PLILALSNPTSQSECTAEEAY WSEGRA
Sbjct: 34 QVIKPTVLIGSSGVGRTFTKEVVEAMTSNNDKPLILALSNPTSQSECTAEEAYQWSEGRA 213
Query: 468 IFASGSPFDPVEYKGKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHDDMLLAASEALAKQV 527
IFASGSPFDPVEYKGK Y SGQ+NNAYIFPGFGLG+V+SGAIRVHDDMLLAASE+LAKQV
Sbjct: 214 IFASGSPFDPVEYKGKVYASGQANNAYIFPGFGLGLVISGAIRVHDDMLLAASESLAKQV 393
Query: 528 TEENYKKGLTYPPFSDIRKISANIAANVAAKAYEL 562
+EENYK GL YPPFS+IR+ISANIAANVAAKAYEL
Sbjct: 394 SEENYKNGLIYPPFSNIRRISANIAANVAAKAYEL 498
Score = 22.7 bits (47), Expect(2) = 1e-73
Identities = 10/25 (40%), Positives = 14/25 (56%)
Frame = +3
Query: 573 QFTIDSEAFGRIKDVVGVWCPFMLV 597
Q ++ A G KDV VWC +L+
Sbjct: 576 QL*VNEAASGSNKDVFRVWCLIVLL 650
>TC204551 homologue to UP|Q9M4Q9 (Q9M4Q9) NADP-dependent malic protein ,
partial (30%)
Length = 856
Score = 263 bits (673), Expect = 1e-70
Identities = 129/165 (78%), Positives = 147/165 (88%)
Frame = +2
Query: 398 EPVSTLLDAVKIIKPTVLIGSSGVGKTFTKEVVEAMTEINKIPLILALSNPTSQSECTAE 457
EPV LLDAV IKPT LIG+SG G+TFTKEV+EAM INK P+IL+LSNPTSQSECTAE
Sbjct: 2 EPVKNLLDAVNKIKPTELIGTSGQGRTFTKEVIEAMASINKRPIILSLSNPTSQSECTAE 181
Query: 458 EAYTWSEGRAIFASGSPFDPVEYKGKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHDDMLL 517
EAY WS+GRAIFASGSPF PVEY+GK + GQ+NNAYIFPGFGLG++MSG IRVHDD+LL
Sbjct: 182 EAYKWSQGRAIFASGSPFPPVEYEGKVFVPGQANNAYIFPGFGLGLIMSGTIRVHDDLLL 361
Query: 518 AASEALAKQVTEENYKKGLTYPPFSDIRKISANIAANVAAKAYEL 562
AASEALA QV++EN+ KGL YPPF++IRKISA+IAANVAAKAYEL
Sbjct: 362 AASEALAAQVSQENFDKGLIYPPFTNIRKISAHIAANVAAKAYEL 496
>BM093124
Length = 420
Score = 231 bits (589), Expect = 7e-61
Identities = 117/139 (84%), Positives = 128/139 (91%), Gaps = 5/139 (3%)
Frame = +2
Query: 298 TTHLVFNDDIQGTAAVVLAGVVASLKLIGGTLPEHTFLFLGAGEAGTGIAELIALEMSKQ 357
++HLVFNDDIQGTA+VVLAG++ASLKL+GGTL +HTFLFLGAGEAGTGIAELIALE+SKQ
Sbjct: 2 SSHLVFNDDIQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQ 181
Query: 358 TKAPIEESRKKIWLVDSK-----GLIVSSRANSLQHFKKPWAHEHEPVSTLLDAVKIIKP 412
TKAP+E++RKKIWLVDSK GLIVSSR SLQHFKKPWAHEHEPV TLLDAVK IKP
Sbjct: 182 TKAPVEKTRKKIWLVDSKVKMPQGLIVSSRLESLQHFKKPWAHEHEPVKTLLDAVKAIKP 361
Query: 413 TVLIGSSGVGKTFTKEVVE 431
TVLIGSSG GKTFTKEVVE
Sbjct: 362 TVLIGSSGAGKTFTKEVVE 418
>AW596922 similar to GP|8118507|gb| NADP-dependent malic protein {Ricinus
communis}, partial (16%)
Length = 460
Score = 147 bits (371), Expect = 1e-35
Identities = 76/137 (55%), Positives = 96/137 (69%)
Frame = +2
Query: 432 AMTEINKIPLILALSNPTSQSECTAEEAYTWSEGRAIFASGSPFDPVEYKGKTYYSGQSN 491
A IN+ P+IL+LSNPT QSECTAEEA WS GRAIFASGSPF PVEY+GK + GQ+N
Sbjct: 2 ARASINERPIILSLSNPT*QSECTAEEANEWSHGRAIFASGSPFPPVEYEGKVFVPGQAN 181
Query: 492 NAYIFPGFGLGIVMSGAIRVHDDMLLAASEALAKQVTEENYKKGLTYPPFSDIRKISANI 551
NAYIFPGFGLG++MSG I V +D + AA EA A Q+++E + L K A++
Sbjct: 182 NAYIFPGFGLGLIMSGTILVQEDQVEAACEA*AAQLSQETMDQELICRTSKQDEKDVAHV 361
Query: 552 AANVAAKAYELVCTATI 568
+AN A+AY+L T +
Sbjct: 362 SANGTAEAYDLGLTTEL 412
>AW132643 similar to GP|8118507|gb| NADP-dependent malic protein {Ricinus
communis}, partial (17%)
Length = 381
Score = 139 bits (350), Expect = 4e-33
Identities = 67/85 (78%), Positives = 76/85 (88%)
Frame = +3
Query: 478 VEYKGKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHDDMLLAASEALAKQVTEENYKKGLT 537
VEY GK + GQ+NNAYIFPGFGLG++MSG IRVHDD+LLAASEALA QVT+++Y KGL
Sbjct: 6 VEYDGKVFVPGQANNAYIFPGFGLGLIMSGTIRVHDDLLLAASEALASQVTQKDYDKGLI 185
Query: 538 YPPFSDIRKISANIAANVAAKAYEL 562
YPPFS+IRKISA IAANVAAKAYEL
Sbjct: 186 YPPFSNIRKISARIAANVAAKAYEL 260
>BQ453046 similar to GP|8894554|emb NAD-dependent malic enzyme (malate
oxidoreductase) {Cicer arietinum}, partial (44%)
Length = 407
Score = 64.7 bits (156), Expect(2) = 2e-22
Identities = 28/58 (48%), Positives = 42/58 (72%)
Frame = +1
Query: 482 GKTYYSGQSNNAYIFPGFGLGIVMSGAIRVHDDMLLAASEALAKQVTEENYKKGLTYP 539
G+ ++ Q+NN Y+FPG GLG ++SGA + D ML AA+E LA +T+E+ +KG+ YP
Sbjct: 223 GEFFHVNQANNMYLFPGIGLGTLLSGARHITDGMLRAAAECLASYMTDEDVQKGILYP 396
Score = 60.1 bits (144), Expect(2) = 2e-22
Identities = 32/65 (49%), Positives = 43/65 (65%), Gaps = 1/65 (1%)
Frame = +2
Query: 410 IKPTVLIGSSGVGKTFTKEVVEAMTE-INKIPLILALSNPTSQSECTAEEAYTWSEGRAI 468
+KP VL+G SGVG F EV++AM E ++ P I A+SNPT +ECTA EA++ +
Sbjct: 2 VKPHVLLGLSGVGGVFNAEVLKAMRESVSTKPAIFAMSNPTMNAECTAIEAFSHAGENIA 181
Query: 469 FASGS 473
FA GS
Sbjct: 182 FAIGS 196
>TC220461 similar to UP|Q9LEN2 (Q9LEN2) NAD-dependent malic enzyme (Malate
oxidoreductase) (Fragment) , partial (59%)
Length = 1101
Score = 96.7 bits (239), Expect = 3e-20
Identities = 51/123 (41%), Positives = 76/123 (61%), Gaps = 1/123 (0%)
Frame = +1
Query: 452 SECTAEEAYTWSEGRAIFASGSPFDPVEY-KGKTYYSGQSNNAYIFPGFGLGIVMSGAIR 510
+ECTA EA++ + +FASGSPF+ V+ G+ + Q+NN Y+FPG GLG ++SGA
Sbjct: 352 AECTAIEAFSHAGENIVFASGSPFENVDLGNGEVGHVNQANNMYLFPGIGLGTLLSGARH 531
Query: 511 VHDDMLLAASEALAKQVTEENYKKGLTYPPFSDIRKISANIAANVAAKAYELVCTATIGE 570
+ D ML AA+E LA +TE++ +KG+ YP IR ++A + A V VC A +
Sbjct: 532 ITDGMLRAAAECLASYMTEDDVRKGILYPSIDCIRDVTAEVGAAV-------VCAAVAEK 690
Query: 571 QGQ 573
Q +
Sbjct: 691 QAE 699
>BG652295 similar to SP|P51615|MAOX_ NADP-dependent malic enzyme (EC
1.1.1.40) (NADP-ME). [Grape] {Vitis vinifera}, partial
(8%)
Length = 410
Score = 82.4 bits (202), Expect = 5e-16
Identities = 42/53 (79%), Positives = 44/53 (82%)
Frame = +2
Query: 63 KGLAFTEKERDAHYLRGLLPPTVSSQQLQEKKLMHNIRQYEVPLQKYVAMMDL 115
K LAF KERDAH L GLLPPT SSQQLQE KLM+NI Y+VPLQKYVAMMDL
Sbjct: 164 KELAFLTKERDAHSLCGLLPPTFSSQQLQENKLMNNISLYQVPLQKYVAMMDL 322
>BG881361 homologue to GP|8894554|emb NAD-dependent malic enzyme (malate
oxidoreductase) {Cicer arietinum}, partial (44%)
Length = 412
Score = 77.0 bits (188), Expect = 2e-14
Identities = 44/112 (39%), Positives = 70/112 (62%), Gaps = 7/112 (6%)
Frame = +2
Query: 375 KGLIVSSRANSLQHFKKPWAHEHEPV------STLLDAVKIIKPTVLIGSSGVGKTFTKE 428
+GL+ + R NSL P+A + +++++ VK I+P VL+G SGVG F +E
Sbjct: 65 EGLVTTER-NSLDPAAAPFAKNPRDIEGLTEGASIIEVVKKIRPHVLLGLSGVGGIFNEE 241
Query: 429 VVEAMTE-INKIPLILALSNPTSQSECTAEEAYTWSEGRAIFASGSPFDPVE 479
V++AM E ++ P I A+SNPT +ECT+ +A+ + +FASGSPF+ V+
Sbjct: 242 VLKAMRESVSTKPAIFAMSNPTMNAECTSIDAFKHAGENIVFASGSPFENVD 397
>BE803412
Length = 411
Score = 68.9 bits (167), Expect = 6e-12
Identities = 33/43 (76%), Positives = 40/43 (92%)
Frame = +2
Query: 520 SEALAKQVTEENYKKGLTYPPFSDIRKISANIAANVAAKAYEL 562
+EALA QV++EN+ KGL YPPF++IRKISA+IAANVAAKAYEL
Sbjct: 242 AEALAAQVSQENFDKGLIYPPFTNIRKISAHIAANVAAKAYEL 370
Score = 57.0 bits (136), Expect = 2e-08
Identities = 24/31 (77%), Positives = 29/31 (93%)
Frame = +1
Query: 489 QSNNAYIFPGFGLGIVMSGAIRVHDDMLLAA 519
Q+NNAYIFPGFGLG++M G IRVHDD+L+AA
Sbjct: 4 QANNAYIFPGFGLGLIMYGTIRVHDDLLVAA 96
>TC232019 homologue to UP|Q9LEN2 (Q9LEN2) NAD-dependent malic enzyme (Malate
oxidoreductase) (Fragment) , partial (37%)
Length = 616
Score = 64.3 bits (155), Expect = 2e-10
Identities = 32/68 (47%), Positives = 42/68 (61%)
Frame = +2
Query: 494 YIFPGFGLGIVMSGAIRVHDDMLLAASEALAKQVTEENYKKGLTYPPFSDIRKISANIAA 553
Y+FPG GLG ++SGA + D ML AASE LA + EE+ KG+ YP IR ++A + A
Sbjct: 5 YLFPGIGLGTLLSGAHLITDGMLQAASECLASYMAEEDILKGILYPSVDSIRDVTAEVGA 184
Query: 554 NVAAKAYE 561
V A E
Sbjct: 185 AVLRAAVE 208
>BG405865 homologue to GP|8118507|gb|A NADP-dependent malic protein {Ricinus
communis}, partial (11%)
Length = 352
Score = 62.8 bits (151), Expect = 4e-10
Identities = 30/40 (75%), Positives = 37/40 (92%)
Frame = +1
Query: 520 SEALAKQVTEENYKKGLTYPPFSDIRKISANIAANVAAKA 559
+EALA QV++EN+ KGL YPPF++IRKISA+IAANVAAKA
Sbjct: 232 AEALAAQVSQENFDKGLIYPPFTNIRKISAHIAANVAAKA 351
Score = 33.5 bits (75), Expect(2) = 5e-05
Identities = 13/15 (86%), Positives = 15/15 (99%)
Frame = +1
Query: 487 SGQSNNAYIFPGFGL 501
+GQ+NNAYIFPGFGL
Sbjct: 1 AGQANNAYIFPGFGL 45
Score = 32.0 bits (71), Expect(2) = 5e-05
Identities = 13/17 (76%), Positives = 16/17 (93%)
Frame = +3
Query: 503 IVMSGAIRVHDDMLLAA 519
++MSG IRVHDD+LLAA
Sbjct: 48 LIMSGTIRVHDDLLLAA 98
>BE352637 similar to SP|P37225|MAON NAD-dependent malic enzyme 59 kDa isoform
mitochondrial precursor (EC 1.1.1.39) (NAD-ME).
[Potato], partial (18%)
Length = 557
Score = 51.6 bits (122), Expect = 1e-06
Identities = 33/87 (37%), Positives = 43/87 (48%), Gaps = 10/87 (11%)
Frame = +2
Query: 52 GYSLLRDPQYNKGLAFTEKERDAHYLRGLLPPTVSSQQLQEKKLMHNIRQYE-------- 103
G +L DP +NK F ERD LRGLLPP V S + Q + M++ R E
Sbjct: 149 GADILHDPWFNKDTGFPLTERDRLGLRGLLPPXVISFEQQYDRFMNSFRSLENNTQGLPD 328
Query: 104 --VPLQKYVAMMDLQERNERLFYKLLI 128
V Q + D RNE L+Y++LI
Sbjct: 329 KVVSWQNGGS*XDCMTRNETLYYRVLI 409
>TC232827 similar to UP|Q94G60 (Q94G60) Sucrose synthase , partial (8%)
Length = 899
Score = 51.2 bits (121), Expect = 1e-06
Identities = 24/34 (70%), Positives = 29/34 (84%)
Frame = +2
Query: 308 QGTAAVVLAGVVASLKLIGGTLPEHTFLFLGAGE 341
QGT +VVLAG+VA+LKL+GG L +H FLFLGA E
Sbjct: 503 QGTTSVVLAGLVAALKLVGGDLTDHMFLFLGARE 604
Score = 30.8 bits (68), Expect = 1.9
Identities = 14/19 (73%), Positives = 17/19 (88%)
Frame = +2
Query: 341 EAGTGIAELIALEMSKQTK 359
+AGTGIAELIALE SK+ +
Sbjct: 812 QAGTGIAELIALETSKRVR 868
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,636,358
Number of Sequences: 63676
Number of extensions: 312636
Number of successful extensions: 1534
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 1517
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1524
length of query: 623
length of database: 12,639,632
effective HSP length: 103
effective length of query: 520
effective length of database: 6,081,004
effective search space: 3162122080
effective search space used: 3162122080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC137546.2


