

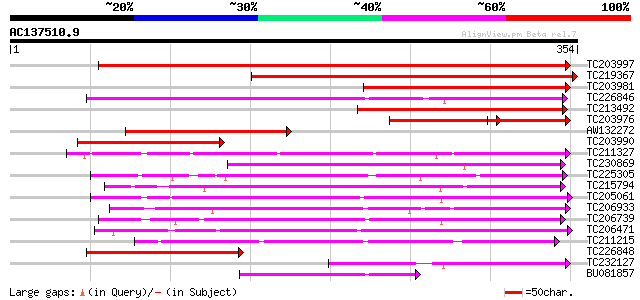
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137510.9 + phase: 0
(354 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC203997 similar to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoenolpyru... 401 e-112
TC219367 weakly similar to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoe... 364 e-101
TC203981 similar to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoenolpyru... 175 3e-44
TC226846 similar to UP|Q9SFE9 (Q9SFE9) T26F17.9 (GONST5 Golgi Nu... 170 8e-43
TC213492 similar to UP|Q9FYE5 (Q9FYE5) Phosphate/phosphoenolpyru... 160 1e-39
TC203976 similar to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoenolpyru... 84 2e-32
AW132272 114 9e-26
TC203990 homologue to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoenolpy... 107 8e-24
TC211327 similar to UP|Q94JT2 (Q94JT2) AT5g17630/K10A8_110, part... 100 1e-21
TC230869 similar to UP|P93390 (P93390) Phosphate/phosphoenolpyru... 92 3e-19
TC225305 similar to UP|CPTR_PEA (P21727) Triose phosphate/phosph... 87 9e-18
TC215794 similar to UP|P93390 (P93390) Phosphate/phosphoenolpyru... 85 6e-17
TC205061 homologue to UP|O64910 (O64910) Glucose-6-phosphate/pho... 84 7e-17
TC206933 similar to UP|Q99322 (Q99322) Mucin (Fragment), partial... 80 1e-15
TC206739 similar to UP|O80688 (O80688) F8K4.1 protein, partial (... 80 2e-15
TC206471 similar to GB|AAR24728.1|38603966|BT010950 At4g09810 {A... 79 2e-15
TC211215 similar to UP|Q9FG70 (Q9FG70) Gb|AAF04433.1, partial (66%) 73 2e-13
TC226848 homologue to UP|Q9SFE9 (Q9SFE9) T26F17.9 (GONST5 Golgi ... 70 1e-12
TC232127 similar to UP|Q94EI9 (Q94EI9) AT3g14410/MLN21_19, parti... 69 3e-12
BU081857 64 8e-11
>TC203997 similar to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoenolpyruvate
translocator protein-like, partial (97%)
Length = 1271
Score = 401 bits (1031), Expect = e-112
Identities = 198/295 (67%), Positives = 244/295 (82%)
Frame = +1
Query: 56 IIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQIHS 115
++++WY SNIGVLLLNKYLLS YG++YPIFLTM HM +C+ +SY AI ++ VP Q I S
Sbjct: 109 LVSAWYSSNIGVLLLNKYLLSNYGFKYPIFLTMCHMTACSLFSYVAIAWLKMVPMQTIRS 288
Query: 116 KKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAEV 175
+ QFLKI ALS IFCFSVV GN SLRYLPVSFNQA+GATTPFFTA+FA+++T K+E
Sbjct: 289 RLQFLKIAALSLIFCFSVVFGNVSLRYLPVSFNQAVGATTPFFTAVFAYVMTFKREAWLT 468
Query: 176 YLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNLL 235
YL L+PVV G+V+++ EP FHLFGF+VC+ +TA RALKSV+QGI+L+SE EKL+SMNLL
Sbjct: 469 YLTLVPVVTGVVIASGGEPSFHLFGFIVCIAATAARALKSVLQGILLSSEGEKLNSMNLL 648
Query: 236 LYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVTKH 295
LYMAP+A + LLP TL +E NV IT+ AR D I++ L+ N+ +AY VNLTNFLVTKH
Sbjct: 649 LYMAPIAVVFLLPATLIMEENVVGITLALARDDVKIIWYLLFNSALAYFVNLTNFLVTKH 828
Query: 296 TSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAKKRSK 350
TSALTLQVLGNAK AVA VVS+LIFRNPV+V GM G+ +T++GVVLYS+AKKRSK
Sbjct: 829 TSALTLQVLGNAKGAVAVVVSILIFRNPVSVTGMMGYSLTVLGVVLYSQAKKRSK 993
>TC219367 weakly similar to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoenolpyruvate
translocator protein-like, partial (65%)
Length = 939
Score = 364 bits (934), Expect = e-101
Identities = 188/203 (92%), Positives = 195/203 (95%)
Frame = +1
Query: 152 GATTPFFTAIFAFLITCKKETAEVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGR 211
GATTPFFTAIFAFLITCKKET EVYLALLPVV GIVV++NSEPLFHLFGFLVCVGSTAGR
Sbjct: 1 GATTPFFTAIFAFLITCKKETGEVYLALLPVVFGIVVASNSEPLFHLFGFLVCVGSTAGR 180
Query: 212 ALKSVVQGIILTSEAEKLHSMNLLLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFI 271
ALKSVVQGI+LTSEAEKLHSMNLLLYMAPLAAMILLP TLYIEGNV A+TIEKA+ DPFI
Sbjct: 181 ALKSVVQGILLTSEAEKLHSMNLLLYMAPLAAMILLPFTLYIEGNVLALTIEKAKGDPFI 360
Query: 272 VFLLIGNATVAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTG 331
VFLL+GNATVAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGM G
Sbjct: 361 VFLLLGNATVAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMAG 540
Query: 332 FGITIMGVVLYSEAKKRSKGASH 354
FGITIMGVVLYSEAKKRSK +H
Sbjct: 541 FGITIMGVVLYSEAKKRSKVTTH 609
>TC203981 similar to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoenolpyruvate
translocator protein-like, partial (42%)
Length = 634
Score = 175 bits (443), Expect = 3e-44
Identities = 89/129 (68%), Positives = 105/129 (80%)
Frame = +3
Query: 222 LTSEAEKLHSMNLLLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATV 281
L SE EKL+SMNLLLYMAP+A + LLP TL +E NV IT+ AR D I++ L+ N+ +
Sbjct: 3 LASEGEKLNSMNLLLYMAPMAVVFLLPATLIMEENVVGITLALARDDSKIIWYLLFNSAL 182
Query: 282 AYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVL 341
AY VNLTNFLVTKHTSALTLQVLGNAK AVA VVS+LIFRNPV+V GM G+ +T+ GV+L
Sbjct: 183 AYFVNLTNFLVTKHTSALTLQVLGNAKGAVAVVVSILIFRNPVSVTGMMGYSLTVFGVIL 362
Query: 342 YSEAKKRSK 350
YSEAKKRSK
Sbjct: 363 YSEAKKRSK 389
>TC226846 similar to UP|Q9SFE9 (Q9SFE9) T26F17.9 (GONST5 Golgi Nucleotide
sugar transporter), partial (97%)
Length = 1334
Score = 170 bits (431), Expect = 8e-43
Identities = 89/303 (29%), Positives = 171/303 (56%), Gaps = 3/303 (0%)
Frame = +2
Query: 49 NLVTTSLIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFV 108
+++ + L I W+ N+ V+++NK++ +++P+ ++ +H + + +Y I +++
Sbjct: 173 SVIRSLLSILQWWAFNVTVIIVNKWIFQKLDFKFPLSVSCVHFICSSIGAYVVIKLLKLK 352
Query: 109 PYQQIHSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITC 168
P + + ++ +IF +S +FC ++V GN SLRY+PVSF Q I + TP T + +L+
Sbjct: 353 PLITVDPEDRWRRIFPMSFVFCINIVLGNVSLRYIPVSFMQTIKSFTPATTVVLQWLVWR 532
Query: 169 KKETAEVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEK 228
K ++ +L+P+V GI++++ +E F++FGF + + K+++ +L K
Sbjct: 533 KYFDWRIWASLVPIVGGILLTSVTELSFNMFGFCAALFGCLATSTKTILAESLL--HGYK 706
Query: 229 LHSMNLLLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPF---IVFLLIGNATVAYLV 285
S+N + YMAP A MIL + +EGN +E + P+ + ++ + +A+ +
Sbjct: 707 FDSINTVYYMAPFATMILALPAMLLEGNGI---LEWLNTHPYPWSALIIIFSSGVLAFCL 877
Query: 286 NLTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEA 345
N + F V T+A+T V GN K AVA +VS LIFRNP++ + G +T++G Y
Sbjct: 878 NFSIFYVIHSTTAVTFNVAGNLKVAVAVLVSWLIFRNPISYLNSVGCTVTLVGCTFYGYV 1057
Query: 346 KKR 348
+ +
Sbjct: 1058RHK 1066
>TC213492 similar to UP|Q9FYE5 (Q9FYE5) Phosphate/phosphoenolpyruvate
translocator-like protein, partial (42%)
Length = 430
Score = 160 bits (404), Expect = 1e-39
Identities = 79/131 (60%), Positives = 101/131 (76%)
Frame = +1
Query: 218 QGIILTSEAEKLHSMNLLLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIG 277
+ I+L+SE EKL+SMNLLLYM+P+A ++LLP L +E NV +T+ A+ + LL
Sbjct: 4 RSILLSSEGEKLNSMNLLLYMSPIAVLVLLPAALIMEPNVVDVTLTLAKDHKSMWLLLFL 183
Query: 278 NATVAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIM 337
N+ +AY NLTNFLVTKHTSALTLQVLGNAK AVA V+S+L+FRNPVTV+GM G+ IT+M
Sbjct: 184 NSVIAYAANLTNFLVTKHTSALTLQVLGNAKGAVAVVISILLFRNPVTVLGMGGYTITVM 363
Query: 338 GVVLYSEAKKR 348
GV Y E K+R
Sbjct: 364 GVAAYGETKRR 396
>TC203976 similar to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoenolpyruvate
translocator protein-like, partial (37%)
Length = 779
Score = 84.3 bits (207), Expect(2) = 2e-32
Identities = 44/70 (62%), Positives = 53/70 (74%)
Frame = +1
Query: 238 MAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVTKHTS 297
MAP+A + LL TL +E NV IT+ AR D I++ L+ N+ +AY VNLTNFLVTKHTS
Sbjct: 1 MAPIAVVXLLXXTLIMEENVVGITLALARDDVKIIWYLLFNSALAYFVNLTNFLVTKHTS 180
Query: 298 ALTLQVLGNA 307
ALTLQVLGNA
Sbjct: 181 ALTLQVLGNA 210
Score = 73.2 bits (178), Expect(2) = 2e-32
Identities = 35/52 (67%), Positives = 42/52 (80%)
Frame = +2
Query: 299 LTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAKKRSK 350
L + LG K AVA VVS+LIFRNPV+V GM G+ +T++GVVLYSEAKKRSK
Sbjct: 185 LPFRYLGTXKGAVAVVVSILIFRNPVSVTGMMGYSLTVLGVVLYSEAKKRSK 340
>AW132272
Length = 313
Score = 114 bits (284), Expect = 9e-26
Identities = 59/104 (56%), Positives = 71/104 (67%)
Frame = +1
Query: 73 YLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQIHSKKQFLKIFALSAIFCFS 132
YLLS YG++YPI LTM HM +C+ SY AI ++ VP Q I S+ QF KI ALS +
Sbjct: 1 YLLSNYGFKYPILLTMCHMTACSLLSYVAIAWMKVVPLQSIRSRVQFFKISALSLVILRF 180
Query: 133 VVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAEVY 176
V GN LRY PVS NQAIGATTPFFT +FA+L+T K+E Y
Sbjct: 181 VGFGNIYLRYPPVSINQAIGATTPFFTDVFAYLMTFKREAWLTY 312
>TC203990 homologue to UP|Q9FFJ6 (Q9FFJ6) Phosphate/phosphoenolpyruvate
translocator protein-like, partial (28%)
Length = 526
Score = 107 bits (267), Expect = 8e-24
Identities = 51/92 (55%), Positives = 70/92 (75%)
Frame = +1
Query: 43 SNINNNNLVTTSLIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAI 102
+ + +++ + T ++++WY SNIGVLLLNKYLLS YG++YPIFLTM HM +C+ +SY AI
Sbjct: 247 TEMKSSSRLFTIGLVSAWYSSNIGVLLLNKYLLSNYGFKYPIFLTMCHMTACSLFSYVAI 426
Query: 103 NVVQFVPYQQIHSKKQFLKIFALSAIFCFSVV 134
++ VP Q I S+ QFLKI ALS +FC SVV
Sbjct: 427 AWLKMVPMQTIRSRLQFLKIAALSLVFCVSVV 522
>TC211327 similar to UP|Q94JT2 (Q94JT2) AT5g17630/K10A8_110, partial (72%)
Length = 1224
Score = 100 bits (248), Expect = 1e-21
Identities = 83/325 (25%), Positives = 150/325 (45%), Gaps = 10/325 (3%)
Frame = +1
Query: 36 SDQRNNGSNI-----NNNNLVTTSLIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLH 90
S+ G N+ N+ NL L+ WYF NI + NK +L+ + + + L
Sbjct: 31 SEANPEGENVAPTEPNSKNL-KLGLVFGLWYFQNIVFNIYNKKVLNIFPFPW---LLASF 198
Query: 91 MLSCAAYSYAAINVVQFVPYQQIHSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQA 150
L + + ++ P +I SK + + + + S + VSF
Sbjct: 199 QLFVGSIWMLVLWSLKLQPCPKI-SKPFIIALLGPALFHTIGHISACVSFSKVAVSFTHV 375
Query: 151 IGATTPFFTAIFAFLITCKKETAEVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAG 210
I + P F+ +F+ ++ K +V+L++LP+VLG ++ +E F++ G + S G
Sbjct: 376 IKSAEPVFSGMFSSVLG-DKYPIQVWLSILPIVLGCSLAAVTEVSFNVQGLWCALISNVG 552
Query: 211 RALKSVVQGIILTSEAEKLHSMNLLLYMAPLAAMILLPVTLYIEGNVFAITIEKA----- 265
L+++ L + E + +NL ++ L+ + L PV +++EG+ + KA
Sbjct: 553 FVLRNIYSKRSLQNFKE-VDGLNLYGWITILSLLYLFPVAIFVEGSQWIPGYYKAIEAIG 729
Query: 266 RSDPFIVFLLIGNATVAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVT 325
++ F ++L+ + +L N +++ S LT V K V V SVL+FRNPV
Sbjct: 730 KASTFYTWVLV-SGVFYHLYNQSSYQALDEISPLTFSVGNTMKRVVVIVSSVLVFRNPVR 906
Query: 326 VMGMTGFGITIMGVVLYSEAKKRSK 350
+ G I I+G LYS+A + K
Sbjct: 907 PLNGLGSAIAILGTFLYSQATSKKK 981
>TC230869 similar to UP|P93390 (P93390) Phosphate/phosphoenolpyruvate
translocator, partial (53%)
Length = 1012
Score = 92.4 bits (228), Expect = 3e-19
Identities = 62/216 (28%), Positives = 112/216 (51%), Gaps = 5/216 (2%)
Frame = +2
Query: 137 NTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAEVYLALLPVVLGIVVSTNSEPLF 196
N SL + VSF I A PFFT + L+ + T V +L+PVV G+ +++ +E F
Sbjct: 5 NISLGKVAVSFTHTIKAMEPFFTVVXXALLLGEMPTFWVVSSLVPVVGGVALASMTEVSF 184
Query: 197 HLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNLLLYMAPLAAMILLPVTLYIEGN 256
+ GF + S ++V+ ++T+E E L ++NL + ++ ++L+P + +EG
Sbjct: 185 NWIGFTTAMASNVTNQSRNVLSKKLMTNEEETLDNINLYSVITIISFLLLVPCAILVEGV 364
Query: 257 VFAIT-IEKARSDPFIVFLLIGNATVA----YLVNLTNFLVTKHTSALTLQVLGNAKAAV 311
F+ + ++ A S V L + +A + ++++ + S +T V K V
Sbjct: 365 KFSPSYLQFAASQGLNVRELCVRSILAAFCYHAYQQVSYMILQMVSPVTHSVGNCVKRVV 544
Query: 312 AAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAKK 347
V SV+ F+ PV+ + G G+ ++GV LYS AK+
Sbjct: 545 VIVSSVIFFQIPVSPVNTLGTGLALVGVFLYSRAKR 652
>TC225305 similar to UP|CPTR_PEA (P21727) Triose phosphate/phosphate
translocator, chloroplast precursor (CTPT) (p36) (E30),
complete
Length = 1707
Score = 87.4 bits (215), Expect = 9e-18
Identities = 80/308 (25%), Positives = 138/308 (43%), Gaps = 10/308 (3%)
Frame = +3
Query: 51 VTTSLIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYA--AINVVQFV 108
+ T +WYF N+ +LNK + +++ Y P F++++H+ AY A+ + +
Sbjct: 384 LVTGFFFFTWYFLNVIFNILNKKIYNYFPY--PYFVSVIHLFVGVAYCLVSWAVGLPKRA 557
Query: 109 PYQQIHSKKQFLKIFALSAIFCFSV--VCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLI 166
P LK+ A+ C ++ V N S + VSF I A PFF A + I
Sbjct: 558 PIDS-----NLLKLLIPVAV-CHALGHVTSNVSFAAVAVSFTHTIKALEPFFNAAASQFI 719
Query: 167 TCKKETAEVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEA 226
+ ++L+L PVV+G+ +++ +E F+ GF+ + S +S+ +T
Sbjct: 720 LGQSIPITLWLSLAPVVIGVSMASLTELSFNWVGFISAMISNISFTYRSIYSKKAMTD-- 893
Query: 227 EKLHSMNLLLYMAPLAAMILLPVTLYIEG-----NVFAITIEKARSDPFIVFLL-IGNAT 280
+ S N+ Y++ +A ++ +P + +EG + F I K F+ L +G
Sbjct: 894 --MDSTNIYAYISIIALIVCIPPAVILEGPTLLKHGFNDAIAKVGLVKFVSDLFWVGMFY 1067
Query: 281 VAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVV 340
Y TN L + + LT V K S+++F N ++ G I I GV
Sbjct: 1068HLYNQVATNTL--ERVAPLTHAVGNVLKRVFVIGFSIIVFGNKISTQTGIGTAIAIAGVA 1241
Query: 341 LYSEAKKR 348
LYS K R
Sbjct: 1242LYSFIKAR 1265
>TC215794 similar to UP|P93390 (P93390) Phosphate/phosphoenolpyruvate
translocator, partial (76%)
Length = 1693
Score = 84.7 bits (208), Expect = 6e-17
Identities = 70/297 (23%), Positives = 132/297 (43%), Gaps = 9/297 (3%)
Frame = +3
Query: 60 WYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQIHSKKQF 119
WY NI + NK +L + YP+ +T++ +V F+ ++ + +
Sbjct: 489 WYLFNIYFNIYNKQVLK--AFHYPVTVTVVQFA-------VGTVLVAFMWGLNLYKRPKL 641
Query: 120 L-----KIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAE 174
I L+A+ + N SL + VSF I A PFF+ I + + + T
Sbjct: 642 SGAMLGAILPLAAVHTLGNLFTNMSLGKVAVSFTHTIKAMEPFFSVILSAMFLGEFPTPW 821
Query: 175 VYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNL 234
V +L+P+V G+ +++ +E F+ GF + S ++V+ + + + + ++ L
Sbjct: 822 VVGSLVPIVGGVALASVTEASFNWAGFWSAMASNVTNQSRNVLSKKAMVKKEDSMDNITL 1001
Query: 235 LLYMAPLAAMILLPVTLYIEGNVFAITIEKARS----DPFIVFLLIGNATVAYLVNLTNF 290
+ ++ +L PV +++EG F ++ +I LL AY ++
Sbjct: 1002FSIITVMSFFLLAPVAIFMEGVKFTPAYLQSAGVNVRQLYIRSLLAALCFHAY--QQVSY 1175
Query: 291 LVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAKK 347
++ + S +T V K V V SV+ F+ PV+ + G I + GV LYS K+
Sbjct: 1176MILQRVSPVTHSVGNCVKRVVVIVSSVIFFQTPVSPVNAFGTAIALAGVFLYSRVKR 1346
>TC205061 homologue to UP|O64910 (O64910)
Glucose-6-phosphate/phosphate-translocator precursor,
partial (89%)
Length = 1898
Score = 84.3 bits (207), Expect = 7e-17
Identities = 70/304 (23%), Positives = 133/304 (43%), Gaps = 3/304 (0%)
Frame = +1
Query: 51 VTTSLIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPY 110
V + A+W+ N+ + NK +L+ Y Y + LT L+C + I+ +
Sbjct: 655 VKIGIYFATWWALNVVFNIYNKKVLNAYPYPW---LTSTLSLACGSLMML-ISWATGIAE 822
Query: 111 QQIHSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKK 170
+ + +F ++ V S+ + VSF I + P F+ + + + +
Sbjct: 823 APKTDPEFWKSLFPVAVAHTIGHVAATVSMSKVAVSFTHIIKSGEPAFSVLVSRFLLGES 1002
Query: 171 ETAEVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLH 230
VYL+L+P++ G ++ +E F++ GF+ + S +++ + + +
Sbjct: 1003FPVPVYLSLIPIIGGCALAAVTELNFNMIGFMGAMISNLAFVFRNIFSK--KGMKGKSVS 1176
Query: 231 SMNLLLYMAPLAAMILLPVTLYIEG-NVFAITIEKARSD--PFIVFLLIGNATVAYLVNL 287
MN ++ L+ IL P + +EG ++A + A S P ++ L + +L N
Sbjct: 1177GMNYYACLSILSLAILTPFAIAVEGPQMWAAGWQTAMSQIGPQFIWWLAAQSVFYHLYNQ 1356
Query: 288 TNFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAKK 347
+++ S LT + K V S++IF PV + G I I+G LYS+AK
Sbjct: 1357VSYMSLDQISPLTFSIGNTMKRISVIVSSIIIFHTPVQPINALGAAIAILGTFLYSQAKL 1536
Query: 348 RSKG 351
S G
Sbjct: 1537*SWG 1548
>TC206933 similar to UP|Q99322 (Q99322) Mucin (Fragment), partial (6%)
Length = 1812
Score = 80.1 bits (196), Expect = 1e-15
Identities = 73/295 (24%), Positives = 137/295 (45%), Gaps = 7/295 (2%)
Frame = +3
Query: 63 SNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQIHSKKQFLKI 122
S++G+++ NK L+S GY + S + +A +V V +S + + +
Sbjct: 357 SSVGIIMANKQLMSNNGYAFSF------ASSLTGFHFAVTALVGLVSNATGYSASKHVPM 518
Query: 123 FAL---SAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKETAEVYLAL 179
+ L S + S+ N SL V F Q + + +++ K + EV +++
Sbjct: 519 WELIWFSLVANMSITGMNFSLMLNSVGFYQISKLSMIPVVCVMEWILHNKHYSREVKMSV 698
Query: 180 LPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNLLLYMA 239
+ VV+G+ V T ++ +L GF+ + +L+ + G + + + S LL A
Sbjct: 699 VVVVIGVGVCTVTDVKVNLKGFMCACIAVLSTSLQQISIGSL--QKKYSIGSFELLSKTA 872
Query: 240 PLAAMILLP----VTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVTKH 295
P+ A+ LL V Y+ G + IT K S + LL + ++A N++ +L
Sbjct: 873 PIQALFLLILGPFVDYYLSGKL--ITNYKMSSGAILCILL--SCSLAVFCNVSQYLCIGR 1040
Query: 296 TSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAKKRSK 350
SA++ QVLG+ K + L+F + +T + G I ++G+V+YS A + K
Sbjct: 1041FSAVSFQVLGHMKTVCVLTLGWLLFDSELTFKNIMGMIIAVVGMVIYSWAVELEK 1205
>TC206739 similar to UP|O80688 (O80688) F8K4.1 protein, partial (77%)
Length = 1668
Score = 79.7 bits (195), Expect = 2e-15
Identities = 71/299 (23%), Positives = 132/299 (43%), Gaps = 7/299 (2%)
Frame = +1
Query: 56 IIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAI----NVVQFVPYQ 111
+ A+W+ N+ + NK +L+ + Y + + LS AA S + V VP
Sbjct: 505 VFATWWALNVVFNIYNKKVLNAFPYPW-----LTSTLSLAAGSLMMLVSWATRVAEVPKV 669
Query: 112 QIHSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCKKE 171
+ K +F ++ V S+ + VSF I + P F+ + + + +
Sbjct: 670 NLDFWKA---LFPVAVAHTIGHVAATVSMSKVAVSFTHIIKSGEPAFSVLVSRFLLGEAF 840
Query: 172 TAEVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHS 231
VYL+LLP++ G ++ +E F++ GF+ + S +++ + + +
Sbjct: 841 PMPVYLSLLPIIGGCALAAVTELNFNMIGFMGAMISNLAFVFRNIFSKKGM--KGMSVSG 1014
Query: 232 MNLLLYMAPLAAMILLPVTLYIEG-NVFAITIEKARSD--PFIVFLLIGNATVAYLVNLT 288
MN ++ ++ +IL P + +EG V+ + A S P V+ + + +L N
Sbjct: 1015MNYYACLSIMSLLILTPFAIAVEGPKVWIAGWQTAVSQIGPNFVWWVAAQSVFYHLYNQV 1194
Query: 289 NFLVTKHTSALTLQVLGNAKAAVAAVVSVLIFRNPVTVMGMTGFGITIMGVVLYSEAKK 347
+++ S LT + K V S+LIF PV + G I I+G LYS+AK+
Sbjct: 1195SYMSLDQISPLTFSIGNTMKRISVIVSSILIFHTPVQPINALGAAIAILGTFLYSQAKQ 1371
>TC206471 similar to GB|AAR24728.1|38603966|BT010950 At4g09810 {Arabidopsis
thaliana;} , partial (94%)
Length = 1533
Score = 79.3 bits (194), Expect = 2e-15
Identities = 74/303 (24%), Positives = 139/303 (45%), Gaps = 5/303 (1%)
Frame = +1
Query: 54 SLIIASWYFS---NIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFVPY 110
S+ ASW F+ ++G++L+NK L++ YG+ + LT LH + + +++ + Y
Sbjct: 268 SIDAASWLFNVVTSVGIILVNKALMATYGFSFATTLTGLHFATTTLLTL----ILKSLGY 435
Query: 111 -QQIHSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVSFNQAIGATTPFFTAIFAFLITCK 169
Q H + F L A F S+V N SL + V F Q + + ++
Sbjct: 436 IQTSHLPVSDIIKFVLFANF--SIVGMNVSLMWNSVGFYQIAKLSMIPVSCFLEVVLDNV 609
Query: 170 KETAEVYLALLPVVLGIVVSTNSEPLFHLFGFLVCVGSTAGRALKSVVQGIILTSEAEKL 229
+ + + L+++ V+LG+ V T ++ + GF+ V + AL+ + +
Sbjct: 610 RYSRDTKLSIVLVLLGVAVCTVTDVSVNAKGFIAAVIAVWSTALQQYY--VHFLQRKYSI 783
Query: 230 HSMNLLLYMAPLAAMILLPVTLYIEGNVFAITIEKARSDPFIVFLLIGNATVAYLVNLTN 289
S NLL + AP A LL V +++ + ++ +I + T+A NL+
Sbjct: 784 GSFNLLGHTAPAQAASLLLVGPFMDYWLTGKRVDAYGYGLTSTLFIILSCTIAVGTNLSQ 963
Query: 290 FLVTKHTSALTLQVLGNAKAAVAAVVSVLIF-RNPVTVMGMTGFGITIMGVVLYSEAKKR 348
F+ +A+T QVLG+ K + ++ + F + + + + G I I G+V Y A +
Sbjct: 964 FICIGRFTAVTFQVLGHMKTILVLILGFIFFGKEGLNLHVVLGMIIAIAGMVWYGNASSK 1143
Query: 349 SKG 351
G
Sbjct: 1144PGG 1152
>TC211215 similar to UP|Q9FG70 (Q9FG70) Gb|AAF04433.1, partial (66%)
Length = 861
Score = 73.2 bits (178), Expect = 2e-13
Identities = 70/268 (26%), Positives = 120/268 (44%), Gaps = 3/268 (1%)
Frame = +1
Query: 79 GYRYPIFLTMLHMLSCAAYSYAAINVVQFVPYQQIHSKKQFLKIFALSAIFCFSVVCGNT 138
G+ +PIFLT +H ++ A A + +P F +FAL + F+ NT
Sbjct: 4 GFNFPIFLTFVHYIT-AWLLLAIFKTLSVLPVSPPSKTTPFSSLFALGVVMAFASGLANT 180
Query: 139 SLRYLPVSFNQ-AIGATTPFFTAIFA-FLITCKKETAEVYLALLPVVLGIVVSTNSEPLF 196
SL+Y V F Q A A TP T + A F+ K + LAL V G+ V+T ++ F
Sbjct: 181 SLKYNSVGFYQMAKIAVTP--TIVLAEFIHFGKTIDFKKVLALAVVSAGVAVATVTDLEF 354
Query: 197 HLFGFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNLLLYMAPLAAMILLPVTLYIEG- 255
+LFG L+ + A+ ++ + + ++ L+ P+ L + +I+
Sbjct: 355 NLFGALIAIAWIIPSAINKILWSTL--QQQGNWTALALMWKTTPITVFFLGALMPWIDPP 528
Query: 256 NVFAITIEKARSDPFIVFLLIGNATVAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAVV 315
V + + S +V L+G +L+ + L TSA T VLG K V +
Sbjct: 529 GVLSFKWDVNNSTAVLVSALLG-----FLLQWSGALALGATSATTHVVLGQFKTCVILLG 693
Query: 316 SVLIFRNPVTVMGMTGFGITIMGVVLYS 343
L+F + V+ + G + + G+ +Y+
Sbjct: 694 GYLLFDSDPGVVSIGGAVVALSGMSVYT 777
>TC226848 homologue to UP|Q9SFE9 (Q9SFE9) T26F17.9 (GONST5 Golgi Nucleotide
sugar transporter), partial (29%)
Length = 463
Score = 70.1 bits (170), Expect = 1e-12
Identities = 27/98 (27%), Positives = 61/98 (61%)
Frame = +3
Query: 49 NLVTTSLIIASWYFSNIGVLLLNKYLLSFYGYRYPIFLTMLHMLSCAAYSYAAINVVQFV 108
+++ + L I W+ N+ V+++NK++ +++P+ ++ +H + + +Y I +++
Sbjct: 168 SVIRSLLSILQWWAFNVTVIIVNKWIFQKLDFKFPLSVSCVHFICSSIGAYVVIKLLKLK 347
Query: 109 PYQQIHSKKQFLKIFALSAIFCFSVVCGNTSLRYLPVS 146
P + + ++ +IF +S +FC ++V GN SLRY+PVS
Sbjct: 348 PLITVDPEDRWRRIFPMSFVFCINIVLGNVSLRYIPVS 461
>TC232127 similar to UP|Q94EI9 (Q94EI9) AT3g14410/MLN21_19, partial (46%)
Length = 820
Score = 68.9 bits (167), Expect = 3e-12
Identities = 45/157 (28%), Positives = 82/157 (51%), Gaps = 6/157 (3%)
Frame = +1
Query: 200 GFLVCVGSTAGRALKSVVQGIILTSEAEKLHSMNLLLYMAPLAAMILLPVTLYIEGNVFA 259
G + +G G AL+ + I + + KL+ ++++ Y++P +A+ L +++E
Sbjct: 34 GVVYQMGGVVGEALRLIFMEIFVKRKGLKLNPISVMYYVSPCSAICLFLPWIFLE----- 198
Query: 260 ITIEKARSDP-----FIVFLLIGNATVAYLVNLTNFLVTKHTSALTLQVLGNAKAAVAAV 314
K + D F LLI N + +NL+ FLV HTSALT++V G K V +
Sbjct: 199 ----KPKMDEHGPWNFPPVLLILNCLCTFALNLSVFLVITHTSALTIRVAGVVKDWVVVL 366
Query: 315 VSVLIFRN-PVTVMGMTGFGITIMGVVLYSEAKKRSK 350
+S ++F + +T++ + G+ I I GV Y+ K + +
Sbjct: 367 LSAVLFADTKLTLINLFGYAIAIAGVAAYNNCKLKKE 477
>BU081857
Length = 400
Score = 64.3 bits (155), Expect = 8e-11
Identities = 38/113 (33%), Positives = 62/113 (54%)
Frame = +2
Query: 144 PVSFNQAIGATTPFFTAIFAFLITCKKETAEVYLALLPVVLGIVVSTNSEPLFHLFGFLV 203
PVSF Q I + TP T + +L+ K ++ +L+P+V GI++++ +E F+ FGF
Sbjct: 2 PVSFMQTIKSFTPATTVVLQWLVWRKYFDWRIWASLIPIVGGILLTSVTELSFNAFGFCA 181
Query: 204 CVGSTAGRALKSVVQGIILTSEAEKLHSMNLLLYMAPLAAMILLPVTLYIEGN 256
+ + K+++ +L K S+N + YMAP A MIL L +EGN
Sbjct: 182 ALLGCLATSTKTILAESLL--HGYKFDSINTVYYMAPFATMILAIPALLLEGN 334
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,541,923
Number of Sequences: 63676
Number of extensions: 204926
Number of successful extensions: 1744
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 1724
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1731
length of query: 354
length of database: 12,639,632
effective HSP length: 98
effective length of query: 256
effective length of database: 6,399,384
effective search space: 1638242304
effective search space used: 1638242304
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC137510.9


