

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137079.8 + phase: 0
(518 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
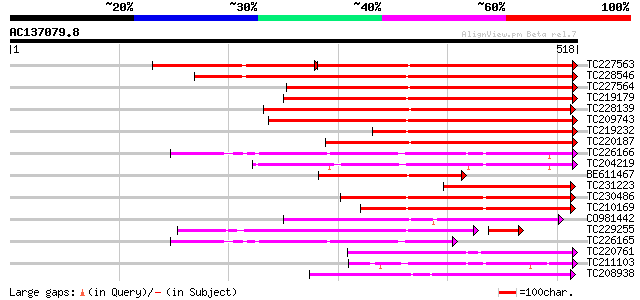
Score E
Sequences producing significant alignments: (bits) Value
TC227563 similar to PIR|E96542|E96542 scarecrow-like protein [im... 394 e-174
TC228546 similar to UP|Q8W584 (Q8W584) AT4g17230/dl4650c, partia... 455 e-128
TC227564 similar to UP|Q9LDL7 (Q9LDL7) SCARECROW gene regulator-... 395 e-110
TC219179 similar to UP|Q9LDL7 (Q9LDL7) SCARECROW gene regulator-... 394 e-110
TC228139 similar to UP|Q8GVE1 (Q8GVE1) Chitin-inducible gibberel... 362 e-100
TC209743 similar to PIR|E86347|E86347 scarecrow-like 1 protein F... 294 7e-80
TC219232 similar to UP|Q9LDL7 (Q9LDL7) SCARECROW gene regulator-... 239 3e-63
TC220187 similar to PIR|E86347|E86347 scarecrow-like 1 protein F... 229 2e-60
TC226166 similar to UP|O23724 (O23724) GAI protein, partial (70%) 218 7e-57
TC204219 similar to UP|Q7Y1B6 (Q7Y1B6) GAI-like protein, partial... 204 1e-52
BE611467 177 1e-44
TC231223 weakly similar to UP|Q8GVE1 (Q8GVE1) Chitin-inducible g... 171 6e-43
TC230486 similar to UP|Q8L553 (Q8L553) SCARECROW transcriptional... 164 7e-41
TC210169 similar to UP|Q8L553 (Q8L553) SCARECROW transcriptional... 153 2e-37
CO981442 146 2e-35
TC229255 similar to UP|Q7X9T5 (Q7X9T5) SCARECROW-like protein, p... 120 2e-33
TC226165 GAI1 [Glycine max] 139 2e-33
TC220761 similar to UP|Q7G7J6 (Q7G7J6) Gibberellin-insensitive p... 131 8e-31
TC211103 weakly similar to UP|Q6S5L6 (Q6S5L6) GAI protein, parti... 131 8e-31
TC208938 weakly similar to UP|Q7X9T5 (Q7X9T5) SCARECROW-like pro... 127 9e-30
>TC227563 similar to PIR|E96542|E96542 scarecrow-like protein [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(71%)
Length = 1274
Score = 394 bits (1012), Expect(2) = e-174
Identities = 193/237 (81%), Positives = 214/237 (89%)
Frame = +2
Query: 282 PPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELR 341
PPKIRITGFDDSTSAYAR GGL IVG RLS+LA+SYNV FEFHAI +P+EV L+DL L+
Sbjct: 449 PPKIRITGFDDSTSAYAREGGLEIVGARLSRLAQSYNVPFEFHAIRAAPTEVELKDLALQ 628
Query: 342 RGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFAR 401
GEAIAVNFAMMLHHVPDE V +NHRDRLVRLAKCLSPK+VTLVEQES+TN LPFF R
Sbjct: 629 PGEAIAVNFAMMLHHVPDECVDS-RNHRDRLVRLAKCLSPKIVTLVEQESHTNNLPFFPR 805
Query: 402 FVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWR 461
FVETMNYY A+FESIDVALPREH+ERINVEQHCLAREVVNL+ACEGAERVERHE+LKKWR
Sbjct: 806 FVETMNYYLAIFESIDVALPREHKERINVEQHCLAREVVNLIACEGAERVERHELLKKWR 985
Query: 462 SCFTMAGFTPYPLSSYINYSIQNLLENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
S FTMAGFTPYPL+S++ SI+NL ++YQGHYTL+E+DGAL LGWMNQ LITS AWR
Sbjct: 986 SRFTMAGFTPYPLNSFVTCSIKNLQQSYQGHYTLEERDGALCLGWMNQVLITSCAWR 1156
Score = 238 bits (606), Expect(2) = e-174
Identities = 117/153 (76%), Positives = 131/153 (85%)
Frame = +3
Query: 131 KWNKMIEMISRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLE 190
+W +M+EMISRGDLKE+L TCAKA++ NDMET EWLMSEL KMVSVSGNPIQRLGAYMLE
Sbjct: 3 RWKRMMEMISRGDLKEMLCTCAKAVAGNDMETTEWLMSELRKMVSVSGNPIQRLGAYMLE 182
Query: 191 ALVARIASSGSIIYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALK 250
ALVAR+ASSGS IYK LKCKEP T ELLSHMH+LYEICPYLKFGYMSANG IAE +K
Sbjct: 183 ALVARLASSGSTIYKVLKCKEP---TGSELLSHMHLLYEICPYLKFGYMSANGAIAEVMK 353
Query: 251 DESEIHIIDFQINQGIQWMSLIQALAGKPGGPP 283
+ESE+HIIDFQINQGIQW+SLI + + P
Sbjct: 354 EESEVHIIDFQINQGIQWVSLIPSCCWQTWSTP 452
>TC228546 similar to UP|Q8W584 (Q8W584) AT4g17230/dl4650c, partial (60%)
Length = 1493
Score = 455 bits (1171), Expect = e-128
Identities = 223/349 (63%), Positives = 279/349 (79%)
Frame = +3
Query: 170 LSKMVSVSGNPIQRLGAYMLEALVARIASSGSIIYKSLKCKEPITATSKELLSHMHVLYE 229
L+KMVSV G+PIQRLGAYMLE L AR+ SSGSIIYK+LKC++P TS +L+++MH+LY+
Sbjct: 30 LAKMVSVGGDPIQRLGAYMLEGLRARLESSGSIIYKALKCEQP---TSNDLMTYMHILYQ 200
Query: 230 ICPYLKFGYMSANGVIAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITG 289
ICPY KF Y SAN VI EA+ +ES I IIDFQI QG QW+ LIQALA +PGGPP + +TG
Sbjct: 201 ICPYWKFAYTSANAVIGEAMLNESRIRIIDFQIAQGTQWLLLIQALASRPGGPPFVHVTG 380
Query: 290 FDDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVN 349
DDS S +ARGGGL IVG+RLS A+S V FEFH+ + SE+ LE+L ++ GEA+ VN
Sbjct: 381 VDDSQSFHARGGGLHIVGKRLSDYAKSCGVPFEFHSAAMCGSELELENLVIQPGEALVVN 560
Query: 350 FAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYY 409
F +LHH+PDE V +NHRDRL+RL K LSPKVVTLVEQESNTN PFF RFVET++YY
Sbjct: 561 FPFVLHHMPDESV-STENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYY 737
Query: 410 FAVFESIDVALPREHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGF 469
A+FESIDVALPR+ ++RIN EQHC+AR++VN+VACEG ER+ERHE+L KWRS F+MAGF
Sbjct: 738 TAMFESIDVALPRDDKQRINAEQHCVARDIVNMVACEGDERLERHELLGKWRSRFSMAGF 917
Query: 470 TPYPLSSYINYSIQNLLENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
P PLSS + +++N+L + +Y LQ +DGALYLGW ++ + TSSAWR
Sbjct: 918 APCPLSSSVTAAVRNMLNEFNENYRLQHRDGALYLGWKSRAMCTSSAWR 1064
>TC227564 similar to UP|Q9LDL7 (Q9LDL7) SCARECROW gene regulator-like
(Phytochrome A signal transduction 1 protein), partial
(53%)
Length = 1107
Score = 395 bits (1014), Expect = e-110
Identities = 196/265 (73%), Positives = 224/265 (83%)
Frame = +3
Query: 254 EIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKL 313
E+HI+DFQI QG QW+SLIQALA +P GPPKIRI+G DDS SAYAR GGL IVG+RLS L
Sbjct: 3 EVHIVDFQIGQGTQWVSLIQALARRPVGPPKIRISGVDDSYSAYARRGGLDIVGKRLSAL 182
Query: 314 AESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLV 373
A+S +V FEF+A+ V +EV+LEDLELR EA+AVNFA+ LHHVPDE V+ NHRDRL+
Sbjct: 183 AQSCHVPFEFNAVRVPVTEVQLEDLELRPYEAVAVNFAISLHHVPDESVNS-HNHRDRLL 359
Query: 374 RLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQH 433
RLAK LSPKVVTLVEQE +TN PF RFVETMNYY AVFESID LPREH+ERINVEQH
Sbjct: 360 RLAKQLSPKVVTLVEQEFSTNNAPFLQRFVETMNYYLAVFESIDTVLPREHKERINVEQH 539
Query: 434 CLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQGHY 493
CLAREVVNL+ACEG ERVERHE+L KWR FT AGFTPYPLSS IN SI++LL++Y GHY
Sbjct: 540 CLAREVVNLIACEGEERVERHELLNKWRMRFTKAGFTPYPLSSVINSSIKDLLQSYHGHY 719
Query: 494 TLQEKDGALYLGWMNQPLITSSAWR 518
TL+E+DGAL+LGWMNQ L+ S AWR
Sbjct: 720 TLEERDGALFLGWMNQVLVASCAWR 794
>TC219179 similar to UP|Q9LDL7 (Q9LDL7) SCARECROW gene regulator-like
(Phytochrome A signal transduction 1 protein), partial
(52%)
Length = 1025
Score = 394 bits (1011), Expect = e-110
Identities = 190/268 (70%), Positives = 221/268 (81%)
Frame = +3
Query: 251 DESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERL 310
DE +HIIDFQI QG QW++LIQA A +PGGPP IRITG DDSTSAYARGGGL IVG RL
Sbjct: 3 DEDRVHIIDFQIGQGSQWITLIQAFAARPGGPPHIRITGIDDSTSAYARGGGLHIVGRRL 182
Query: 311 SKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRD 370
SKLAE + V FEFHA +S +V+L +L +R GEA+AVNFA MLHH+PDE V +NHRD
Sbjct: 183 SKLAEHFKVPFEFHAAAISGFDVQLHNLGVRPGEALAVNFAFMLHHMPDESV-STQNHRD 359
Query: 371 RLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINV 430
RL+RL + LSPKVVTLVEQESNTN FF RF+ET+NYY A+FESIDV LPREH+ERINV
Sbjct: 360 RLLRLVRSLSPKVVTLVEQESNTNTAAFFPRFLETLNYYTAMFESIDVTLPREHKERINV 539
Query: 431 EQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQ 490
EQHCLAR++VN++ACEG ERVERHEVL KWRS F MAGFTPYPLSS +N +I+ LLENY
Sbjct: 540 EQHCLARDLVNIIACEGVERVERHEVLGKWRSRFAMAGFTPYPLSSLVNGTIKKLLENYS 719
Query: 491 GHYTLQEKDGALYLGWMNQPLITSSAWR 518
Y L+E+DGALYLGWMN+ L+ S AW+
Sbjct: 720 DRYRLEERDGALYLGWMNRDLVASCAWK 803
>TC228139 similar to UP|Q8GVE1 (Q8GVE1) Chitin-inducible
gibberellin-responsive protein, partial (47%)
Length = 1497
Score = 362 bits (930), Expect = e-100
Identities = 175/285 (61%), Positives = 217/285 (75%)
Frame = +1
Query: 233 YLKFGYMSANGVIAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDD 292
YLKFGYM+ANG IA+A ++E IHIIDFQI QG QWM+L+QALA +PGG P +RITG DD
Sbjct: 4 YLKFGYMAANGAIAQACRNEDHIHIIDFQIAQGTQWMTLLQALAARPGGAPHVRITGIDD 183
Query: 293 STSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAM 352
S YARG GL +VG+RL+ ++E + + EFH + V V E L++R GEA+AVNF +
Sbjct: 184 PVSKYARGDGLEVVGKRLALMSEKFGIPVEFHGVPVFAPNVTREMLDIRPGEALAVNFPL 363
Query: 353 MLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAV 412
LHH DE VH N RD L+RL + LSPKV TLVEQESNTN PFF RF+ET++YY A+
Sbjct: 364 QLHHTADESVHVS-NPRDGLLRLVRSLSPKVTTLVEQESNTNTTPFFNRFIETLDYYLAI 540
Query: 413 FESIDVALPREHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPY 472
FESIDV LPR+ +ERINVEQHCLAR++VN++ACEG ERVERHE+ KW+S MAGF
Sbjct: 541 FESIDVTLPRDSKERINVEQHCLARDIVNIIACEGKERVERHELFGKWKSRLKMAGFQQC 720
Query: 473 PLSSYINYSIQNLLENYQGHYTLQEKDGALYLGWMNQPLITSSAW 517
PLSSY+N I++LL Y HYTL EKDGA+ LGW ++ LI++SAW
Sbjct: 721 PLSSYVNSVIRSLLMCYSEHYTLVEKDGAMLLGWKDRNLISASAW 855
>TC209743 similar to PIR|E86347|E86347 scarecrow-like 1 protein F24J8.8 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(47%)
Length = 1078
Score = 294 bits (752), Expect = 7e-80
Identities = 142/283 (50%), Positives = 198/283 (69%), Gaps = 1/283 (0%)
Frame = +1
Query: 237 GYMSANGVIAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSA 296
G + ANG IAE ++DE ++HIIDF I+QG Q+++LIQ LA PG PP +R+T DD S
Sbjct: 1 GRVGANGAIAEVVRDEKKVHIIDFDISQGTQYITLIQTLASMPGRPPHVRLTAVDDPESV 180
Query: 297 YARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHH 356
GG+ I+G+RL KLAE + FEF A+ S V L R GEA+ VNFA LHH
Sbjct: 181 QRSIGGINIIGQRLEKLAEELRLPFEFRAVASRTSNVTQSMLNCRPGEALVVNFAFQLHH 360
Query: 357 VPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESI 416
+ DE V N RD+L+R+ K L+PK+VT+VEQ+ NTN PF RF+ET NYY AVF ++
Sbjct: 361 MRDETV-STVNERDQLLRMVKSLNPKIVTVVEQDMNTNTSPFLPRFIETYNYYSAVFNTL 537
Query: 417 DVALPREHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSS 476
D LPRE ++R+NVE+ CLA+++VN+VACEG ER+ER+EV KWR+ +MAGFTP P+S+
Sbjct: 538 DATLPRESQDRMNVERQCLAKDIVNIVACEGEERIERYEVAGKWRARLSMAGFTPSPMST 717
Query: 477 YINYSIQNL-LENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
+ +I+NL ++ Y + ++E+ G L+ GW ++ LI +SAW+
Sbjct: 718 NVREAIRNLIIKQYCDKFKIKEEMGGLHFGWEDKNLIVASAWK 846
>TC219232 similar to UP|Q9LDL7 (Q9LDL7) SCARECROW gene regulator-like
(Phytochrome A signal transduction 1 protein), partial
(38%)
Length = 881
Score = 239 bits (609), Expect = 3e-63
Identities = 116/187 (62%), Positives = 149/187 (79%)
Frame = +3
Query: 332 EVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQES 391
EV ++E+ GEA+AV+F +LHH+PDE V +NHRDRL+RL K LSPKVVT+VEQES
Sbjct: 9 EVVRGNIEVLPGEALAVSFPYVLHHMPDESV-STENHRDRLLRLVKRLSPKVVTIVEQES 185
Query: 392 NTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVVNLVACEGAERV 451
NTN PFF RFVET++YY A+FESIDVA PR+ ++RI+ EQHC+AR++VN++ACEG ERV
Sbjct: 186 NTNTSPFFHRFVETLDYYTAMFESIDVACPRDDKKRISAEQHCVARDIVNMIACEGVERV 365
Query: 452 ERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQGHYTLQEKDGALYLGWMNQPL 511
ERHE+L KWRS +MAGF LSS + +IQNLL+ + +Y L+ +DGALYLGWMN+ +
Sbjct: 366 ERHELLGKWRSRLSMAGFKQCQLSSSVMVAIQNLLKEFSQNYRLEHRDGALYLGWMNRHM 545
Query: 512 ITSSAWR 518
TSSAWR
Sbjct: 546 ATSSAWR 566
>TC220187 similar to PIR|E86347|E86347 scarecrow-like 1 protein F24J8.8 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(39%)
Length = 810
Score = 229 bits (584), Expect = 2e-60
Identities = 117/231 (50%), Positives = 156/231 (66%), Gaps = 1/231 (0%)
Frame = +3
Query: 289 GFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAV 348
G DD S GGL +G+RL KLAE+ + FEF A+ S V L EA+ V
Sbjct: 3 GVDDPESVQRSVGGLQNIGQRLEKLAEALGLPFEFRAVASRTSIVTPSMLNCSPDEALVV 182
Query: 349 NFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNY 408
NFA LHH+PDE V N RD+L+RL K L+PK+VT+VEQ+ NTN PF RFVE NY
Sbjct: 183 NFAFQLHHMPDESVSTA-NERDQLLRLVKSLNPKLVTVVEQDVNTNTTPFLPRFVEAYNY 359
Query: 409 YFAVFESIDVALPREHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAG 468
Y AVFES+D LPRE ++R+NVE+ CLAR++VN+VACEG +R+ER+EV KWR+ TMAG
Sbjct: 360 YSAVFESLDATLPRESQDRMNVERQCLARDIVNVVACEGEDRIERYEVAGKWRARMTMAG 539
Query: 469 FTPYPLSSYINYSIQNLLEN-YQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
FT P+S+ + I+ L++ Y Y ++E+ GAL+ GW ++ LI +SAW+
Sbjct: 540 FTSSPMSTNVTDEIRKLIKTVYCDRYKIKEEMGALHFGWEDKNLIVASAWK 692
>TC226166 similar to UP|O23724 (O23724) GAI protein, partial (70%)
Length = 1613
Score = 218 bits (554), Expect = 7e-57
Identities = 131/374 (35%), Positives = 207/374 (55%), Gaps = 3/374 (0%)
Frame = +1
Query: 148 LFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSIIYKSL 207
L CA+A+ N++ AE L+ ++ + ++++ Y EAL RI Y+
Sbjct: 124 LMACAEAVENNNLAVAEALVKQIGFLAVSQVGAMRKVAIYFAEALARRI-------YRVF 282
Query: 208 KCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQINQGIQ 267
+ + S L H YE CPYLKF + +AN VI EA + ++ +H+IDF INQG+Q
Sbjct: 283 PLQHSL---SDSLQIHF---YETCPYLKFAHFTANQVILEAFQGKNRVHVIDFGINQGMQ 444
Query: 268 WMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHA-I 326
W +L+QALA + GGPP R+TG A L VG +L++LAE NV FE+ +
Sbjct: 445 WPALMQALAVRTGGPPVFRLTGI--GPPAADNSDHLQEVGWKLAQLAEEINVQFEYRGFV 618
Query: 327 GVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTL 386
S +++ L+LR E++AVN H + ++++ + + + P+++T+
Sbjct: 619 ANSLADLDASMLDLREDESVAVNSVFEFH-----KLLARPGAVEKVLSVVRQIRPEILTV 783
Query: 387 VEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVVNLVACE 446
VEQE+N N L F RF E+++YY +F+S++ P ++ E + L +++ N+VACE
Sbjct: 784 VEQEANHNGLSFVDRFTESLHYYSTLFDSLE-GSPVNPNDKAMSEVY-LGKQICNVVACE 957
Query: 447 GAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQG--HYTLQEKDGALYL 504
G +RVERHE L +WR+ F GF+P L S LL + G Y ++E +G L L
Sbjct: 958 GMDRVERHETLNQWRNRFGSTGFSPVHLGSNAYKQASMLLSLFGGGDGYRVEENNGCLML 1137
Query: 505 GWMNQPLITSSAWR 518
GW +PLI +S W+
Sbjct: 1138GWHTRPLIATSVWQ 1179
>TC204219 similar to UP|Q7Y1B6 (Q7Y1B6) GAI-like protein, partial (50%)
Length = 1208
Score = 204 bits (518), Expect = 1e-52
Identities = 123/308 (39%), Positives = 179/308 (57%), Gaps = 12/308 (3%)
Frame = +1
Query: 223 HMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQINQGIQWMSLIQALAGKPGGP 282
HMH YE CPYLKF + +AN I EA ++H+IDF + QG+QW +L+QALA +PGGP
Sbjct: 7 HMH-FYESCPYLKFAHFTANQAILEAFATAGKVHVIDFGLKQGMQWPALMQALALRPGGP 183
Query: 283 PKIRITGF----DDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHA-IGVSPSEVRLED 337
P R+TG D+T A L VG +L++LA++ V FEF + S +++ +
Sbjct: 184 PTFRLTGIGPPQPDNTDA------LQQVGWKLAQLAQNIGVQFEFRGFVCNSLADLDPKM 345
Query: 338 LELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELP 397
LE+R GEA+AVN LH + D+++ K + PK+VT+VEQE+N N
Sbjct: 346 LEIRPGEAVAVNSVFELHR-----MLARPGSVDKVLDTVKKIKPKIVTIVEQEANHNGPG 510
Query: 398 FFARFVETMNYYFAVFESID-----VALPREHRERINVEQHCLAREVVNLVACEGAERVE 452
F RF E ++YY ++F+S++ L +++ + E + L R++ N+VA EGA+RVE
Sbjct: 511 FLDRFTEALHYYSSLFDSLEGSSSSTGLGSPNQDLLMSELY-LGRQICNVVANEGADRVE 687
Query: 453 RHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQG--HYTLQEKDGALYLGWMNQP 510
RHE L +WR AGF P L S LL + G Y ++E +G L LGW +P
Sbjct: 688 RHETLSQWRGRLDSAGFDPVHLGSNAFKQASMLLALFAGGDGYRVEENNGCLMLGWHTRP 867
Query: 511 LITSSAWR 518
LI +SAW+
Sbjct: 868 LIATSAWK 891
>BE611467
Length = 412
Score = 177 bits (448), Expect = 1e-44
Identities = 95/135 (70%), Positives = 107/135 (78%)
Frame = +3
Query: 283 PKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRR 342
PKIRI+G DDS SAYARGGGL IVG+RLS A+S +V FEF+A+ V S+V+LEDLEL
Sbjct: 3 PKIRISGVDDSYSAYARGGGLDIVGKRLSAHAQSCHVPFEFNAVRVPASQVQLEDLELLP 182
Query: 343 GEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARF 402
EA+AVNFA+ LHHVPDE V+ NHRDRL+RLAK LSPKVVTLVEQE NTN PF RF
Sbjct: 183 YEAVAVNFAISLHHVPDESVN-SHNHRDRLLRLAKRLSPKVVTLVEQEFNTNNAPFLQRF 359
Query: 403 VETMNYYFAVFESID 417
ETM YY AV ESID
Sbjct: 360 DETMKYYLAVXESID 404
>TC231223 weakly similar to UP|Q8GVE1 (Q8GVE1) Chitin-inducible
gibberellin-responsive protein, partial (22%)
Length = 653
Score = 171 bits (434), Expect = 6e-43
Identities = 77/121 (63%), Positives = 100/121 (82%)
Frame = +1
Query: 397 PFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVVNLVACEGAERVERHEV 456
PFF RF+ET++YY A+FESIDV+LPR+ + +IN+EQHCLAR++VN++ACEG ERVERHE+
Sbjct: 13 PFFNRFIETLDYYLAIFESIDVSLPRKSKVQINMEQHCLARDIVNIIACEGKERVERHEL 192
Query: 457 LKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQGHYTLQEKDGALYLGWMNQPLITSSA 516
L KW+S TMAGF YPLSSY+N I++LL Y HY L EKDGA+ LGW ++ LI++SA
Sbjct: 193 LGKWKSRLTMAGFRQYPLSSYMNSVIRSLLRCYSKHYNLVEKDGAMLLGWKDRNLISTSA 372
Query: 517 W 517
W
Sbjct: 373 W 375
>TC230486 similar to UP|Q8L553 (Q8L553) SCARECROW transcriptional
regulator-like, partial (44%)
Length = 886
Score = 164 bits (416), Expect = 7e-41
Identities = 92/215 (42%), Positives = 131/215 (60%)
Frame = +2
Query: 303 LGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDV 362
L IVG L + AE + FEF + +E+ E L E +AVNFA L+ +PDE V
Sbjct: 44 LNIVGVLLGRHAEKLGIGFEFKVLTRRIAELTRESLGCDADEPLAVNFAYKLYRMPDESV 223
Query: 363 HGGKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPR 422
+N RD+L+R K L+P+VVTLVEQ++N N PF AR E YY A+F+S++ + R
Sbjct: 224 -STENPRDKLLRRVKTLAPRVVTLVEQDANANTAPFVARVTELCAYYGALFDSLESTMAR 400
Query: 423 EHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSI 482
E+ +R+ +E+ L+R+VVN VACEG +RVER EV KWR+ +MAGF PLS + SI
Sbjct: 401 ENLKRVRIEEG-LSRKVVNSVACEGRDRVERCEVFGKWRARMSMAGFRLKPLSQRVADSI 577
Query: 483 QNLLENYQGHYTLQEKDGALYLGWMNQPLITSSAW 517
+ L ++ ++G + GWM + L +SAW
Sbjct: 578 KARLGGAGNRVAVKVENGGICFGWMGRTLTVASAW 682
>TC210169 similar to UP|Q8L553 (Q8L553) SCARECROW transcriptional
regulator-like, partial (47%)
Length = 872
Score = 153 bits (386), Expect = 2e-37
Identities = 86/197 (43%), Positives = 120/197 (60%)
Frame = +1
Query: 321 FEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLS 380
FEF + +E+ E L+ EA+AVNFA L+ +PDE V +N RD L+R K L+
Sbjct: 91 FEFKVLIRRIAELTRESLDCDADEALAVNFAYKLYRMPDESV-STENPRDELLRRVKALA 267
Query: 381 PKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVV 440
P+VVTLVEQE+N N PF AR E YY A+F+S++ + RE+ R+ +E+ L+R+V
Sbjct: 268 PRVVTLVEQEANANTAPFVARVSELCAYYGALFDSLESTMARENSARVRIEEG-LSRKVG 444
Query: 441 NLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLENYQGHYTLQEKDG 500
N VACEG RVER EV KWR+ +MAGF PLS + SI+ L ++ ++G
Sbjct: 445 NSVACEGRNRVERCEVFGKWRARMSMAGFRLKPLSQRVAESIKARLGGAGNRVAVKVENG 624
Query: 501 ALYLGWMNQPLITSSAW 517
+ GWM + L +SAW
Sbjct: 625 GICFGWMGRTLTVASAW 675
>CO981442
Length = 791
Score = 146 bits (369), Expect = 2e-35
Identities = 94/261 (36%), Positives = 141/261 (54%), Gaps = 5/261 (1%)
Frame = -2
Query: 251 DESEIHIIDFQINQGIQWMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERL 310
D IHIIDF I G +W +LI L+ + GGPPK+RITG D + G RL
Sbjct: 781 DAKAIHIIDFGIRYGFKWPALISRLSRRSGGPPKLRITGIDVPQPGLRPQERVLETGRRL 602
Query: 311 SKLAESYNVAFEFHAIGVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRD 370
+ + +NV FEF+AI +R+EDL++ E +AVN H+ DE V N RD
Sbjct: 601 ANFCKRFNVPFEFNAIAQRWDTIRVEDLKIEPNEFVAVNCLFQFEHLLDETVVLN-NSRD 425
Query: 371 RLVRLAKCLSPKVVT--LVEQESNTNELPFF-ARFVETMNYYFAVFESIDVALPREHRER 427
++RL K +P + +V + + ++PFF +RF E + +Y A+F+ +D + R+ R
Sbjct: 424 AVLRLIKNANPDIFVHGIV---NGSYDVPFFVSRFREALFHYTALFDMLDTNVARQDPMR 254
Query: 428 INVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQN-LL 486
+ E+ RE+VN++ACEG ERVER + K+W+ GF PL I +++ L
Sbjct: 253 LMFEKELFGREIVNIIACEGFERVERPQTYKQWQLRNMRNGFRLLPLDHRIIGKLKDRLR 74
Query: 487 ENYQGHYTLQEKDGALYL-GW 506
++ + L E DG L GW
Sbjct: 73 DDAHNNNFLLEVDGDWVLQGW 11
>TC229255 similar to UP|Q7X9T5 (Q7X9T5) SCARECROW-like protein, partial (37%)
Length = 1561
Score = 120 bits (302), Expect(2) = 2e-33
Identities = 77/275 (28%), Positives = 135/275 (49%)
Frame = +1
Query: 154 AISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSIIYKSLKCKEPI 213
A++ D A L+S++ + S G+ +QRL Y L R+A+ G+ Y L+
Sbjct: 1 AVASFDQRNANDLLSQIRQHSSAFGDGLQRLAHYFANGLQTRLAA-GTPSYTPLE----- 162
Query: 214 TATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQINQGIQWMSLIQ 273
TS ++L + P + A I +++E +HIIDF I G QW LI+
Sbjct: 163 GTTSADMLKAYKLYVTSSPLQRLTNYLATKTIVSLVENEGSVHIIDFGICYGFQWPCLIK 342
Query: 274 ALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEV 333
L+ + GGPP++RITG + + + G RL+ + + V FE++ + +
Sbjct: 343 KLSERHGGPPRLRITGIELPQPGFRPAERVEETGRRLANYCKKFKVPFEYNCLAQKWETI 522
Query: 334 RLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNT 393
+L DL++ R E V+ L ++PDE V K+ RD +++L + ++P +
Sbjct: 523 KLADLKIDRNEVTVVSCFYRLKNLPDETV-DVKSPRDAVLKLIRRINPNMFIHGVVNGTY 699
Query: 394 NELPFFARFVETMNYYFAVFESIDVALPREHRERI 428
N F RF E + ++ ++F+ + +PRE ER+
Sbjct: 700 NAPFFLTRFREALYHFSSLFDMFEANVPREDPERV 804
Score = 40.4 bits (93), Expect(2) = 2e-33
Identities = 17/32 (53%), Positives = 23/32 (71%)
Frame = +3
Query: 438 EVVNLVACEGAERVERHEVLKKWRSCFTMAGF 469
+ +N++ACEGAERVER E K+W+ AGF
Sbjct: 804 DAINVIACEGAERVERPETYKQWQVRNQRAGF 899
>TC226165 GAI1 [Glycine max]
Length = 1313
Score = 139 bits (351), Expect = 2e-33
Identities = 87/263 (33%), Positives = 142/263 (53%), Gaps = 1/263 (0%)
Frame = +3
Query: 148 LFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALVARIASSGSIIYKSL 207
L CA+A+ N++ AE L+ ++ + ++++ Y EAL RI Y+
Sbjct: 579 LMACAEAVENNNLAVAEALVKQIGFLALSQVGAMRKVATYFAEALARRI-------YRVF 737
Query: 208 KCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDFQINQGIQ 267
+ + S L H YE CPYLKF + +AN I EA + ++ +H+IDF INQG+Q
Sbjct: 738 PQQHSL---SDSLQIHF---YETCPYLKFAHFTANQAILEAFQGKNRVHVIDFGINQGMQ 899
Query: 268 WMSLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHA-I 326
W +L+QALA + GPP R+TG A L VG +L++LAE +V FE+ +
Sbjct: 900 WPALMQALALRNDGPPVFRLTGI--GPPAADNSDHLQEVGWKLAQLAERIHVQFEYRGFV 1073
Query: 327 GVSPSEVRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTL 386
S +++ L+LR E++AVN H + ++++ + + + P+++T+
Sbjct: 1074ANSLADLDASMLDLREDESVAVNSVFEFH-----KLLARPGAVEKVLSVVRQIRPEILTV 1238
Query: 387 VEQESNTNELPFFARFVETMNYY 409
VEQE+N N L F RF E+++YY
Sbjct: 1239VEQEANHNGLSFVDRFTESLHYY 1307
>TC220761 similar to UP|Q7G7J6 (Q7G7J6) Gibberellin-insensitive protein
OsGAI, partial (23%)
Length = 719
Score = 131 bits (329), Expect = 8e-31
Identities = 80/212 (37%), Positives = 119/212 (55%), Gaps = 2/212 (0%)
Frame = +3
Query: 309 RLSKLAESYNVAFEFHAIGVSPSE-VRLEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKN 367
RL++LA S NV F F + E V+ L++ EA+AVN M LH + D +
Sbjct: 12 RLAELAXSVNVRFAFRGVAAXXXEDVKPWMLQVNPNEAVAVNSIMQLHRLLASDSDPIGS 191
Query: 368 HRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREHRER 427
+ ++ + L+PK++++VEQE+N N+ F RF E ++YY VF+S++ A P E +
Sbjct: 192 GIETVLGWIRSLNPKIISVVEQEANHNQDRFLERFTEALHYYSTVFDSLE-ACPVEPDKA 368
Query: 428 INVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPLSSYINYSIQNLLE 487
+ + L RE+ N+V+ EG RVERHE L KWR AGF P L S LL
Sbjct: 369 L--AEMYLQREICNVVSSEGPARVERHEPLAKWRERLEKAGFKPLHLGSNAYKQASMLLT 542
Query: 488 NYQGH-YTLQEKDGALYLGWMNQPLITSSAWR 518
+ Y+++E G L LGW ++PLI +SAW+
Sbjct: 543 LFSAEGYSVEENQGCLTLGWHSRPLIAASAWQ 638
>TC211103 weakly similar to UP|Q6S5L6 (Q6S5L6) GAI protein, partial (27%)
Length = 807
Score = 131 bits (329), Expect = 8e-31
Identities = 86/216 (39%), Positives = 123/216 (56%), Gaps = 7/216 (3%)
Frame = +3
Query: 310 LSKLAESYNVAFEFHAIGVSPSEVRLED-----LELRRGEAIAVNFAMMLHHVPDEDVHG 364
L++LA S NV F F + RLED L++ EA+AVN M LH +
Sbjct: 3 LAELARSVNVRFAFRGVAAW----RLEDVKPWMLQVSPNEAVAVNSIMQLHRLT-----A 155
Query: 365 GKNHRDRLVRLAKCLSPKVVTLVEQESNTNELPFFARFVETMNYYFAVFESIDVALPREH 424
K+ + ++ + L+PK+VT+VEQE+N N F RF E ++YY VF+S+D A P E
Sbjct: 156 VKSAVEEVLGWIRILNPKIVTVVEQEANHNGEGFLERFTEALHYYSTVFDSLD-ACPVEP 332
Query: 425 RERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGFTPYPL--SSYINYSI 482
+ E + L RE+ N+V CEG R+ERHE L KWR AGF P L ++Y S+
Sbjct: 333 DKAALAEMY-LQREICNVVCCEGPARLERHEPLAKWRDRLGKAGFRPLHLGFNAYKQASM 509
Query: 483 QNLLENYQGHYTLQEKDGALYLGWMNQPLITSSAWR 518
L + +G + +QE G+L LGW ++PLI +SAW+
Sbjct: 510 LLTLFSAEG-FCVQENQGSLTLGWHSRPLIAASAWQ 614
>TC208938 weakly similar to UP|Q7X9T5 (Q7X9T5) SCARECROW-like protein,
partial (31%)
Length = 915
Score = 127 bits (320), Expect = 9e-30
Identities = 75/245 (30%), Positives = 127/245 (51%), Gaps = 2/245 (0%)
Frame = +2
Query: 275 LAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVR 334
L+ +PGGPPK+R+ G D + + G L K + + V FE++ + +R
Sbjct: 2 LSERPGGPPKLRMMGIDLPQPGFRPAERVEETGRWLEKYCKRFGVPFEYNCLAQKWETIR 181
Query: 335 LEDLELRRGEAIAVNFAMMLHHVPDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQESNTN 394
LEDL++ R E VN L ++ DE V RD L+RL + ++P + + + T
Sbjct: 182 LEDLKIDRSEVTVVNCLYRLKNLSDETVTANCP-RDALLRLIRRINPNIF-MHGVVNGTY 355
Query: 395 ELPFFA-RFVETMNYYFAVFESIDVALPREHRERINVEQHCLAREVVNLVACEGAERVER 453
PFF RF E + ++ ++F+ + +PRE R+ +E+ R+ +N++ACEGAERVER
Sbjct: 356 NAPFFVTRFREALFHFSSLFDMFEANVPREDPSRLMIEKGLFGRDAINVIACEGAERVER 535
Query: 454 HEVLKKWRSCFTMAGFTPYPLS-SYINYSIQNLLENYQGHYTLQEKDGALYLGWMNQPLI 512
E K+W+ AGF PL+ ++N + + + + + + E + GW + L
Sbjct: 536 PETYKQWQVRNQRAGFKQLPLAPEHVNRVKEMVKKEHHKDFVVDEDGKWVLQGWKGRILF 715
Query: 513 TSSAW 517
S+W
Sbjct: 716 AVSSW 730
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,640,235
Number of Sequences: 63676
Number of extensions: 293420
Number of successful extensions: 1622
Number of sequences better than 10.0: 97
Number of HSP's better than 10.0 without gapping: 1539
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1554
length of query: 518
length of database: 12,639,632
effective HSP length: 102
effective length of query: 416
effective length of database: 6,144,680
effective search space: 2556186880
effective search space used: 2556186880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC137079.8


