

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137077.8 - phase: 0
(1091 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
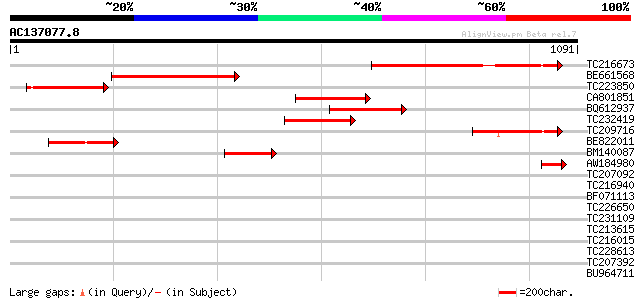
Score E
Sequences producing significant alignments: (bits) Value
TC216673 similar to UP|O23575 (O23575) G2484-1 protein (Fragment... 489 e-138
BE661568 similar to GP|20466528|gb G2484-1 protein {Arabidopsis ... 298 7e-81
TC223850 similar to UP|Q8LPI8 (Q8LPI8) G2484-1 protein (At4g1733... 224 1e-58
CA801851 weakly similar to GP|20466528|gb G2484-1 protein {Arabi... 205 1e-52
BQ612937 similar to GP|20466528|gb G2484-1 protein {Arabidopsis ... 198 9e-51
TC232419 192 5e-49
TC209716 similar to UP|O23575 (O23575) G2484-1 protein (Fragment... 188 1e-47
BE822011 similar to GP|20466528|gb G2484-1 protein {Arabidopsis ... 179 6e-45
BM140087 similar to GP|20466528|gb G2484-1 protein {Arabidopsis ... 164 2e-40
AW184980 78 2e-14
TC207092 similar to UP|Q94ET8 (Q94ET8) LCI5, partial (11%) 42 0.002
TC216940 weakly similar to GB|AAP37755.1|30725466|BT008396 At1g6... 41 0.003
BF071113 41 0.003
TC226650 similar to UP|Q94BZ2 (Q94BZ2) At1g30970/F17F8_14, parti... 40 0.007
TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, par... 40 0.007
TC213615 similar to UP|Q9SUL7 (Q9SUL7) SNF1 like protein kinase ... 39 0.012
TC216015 similar to UP|Q9M610 (Q9M610) Ttg1-like protein, partia... 39 0.016
TC228613 weakly similar to UP|PE31_ARATH (Q9LHA7) Peroxidase 31 ... 38 0.021
TC207392 similar to UP|BNK_DROME (P40794) Bottleneck protein, pa... 38 0.021
BU964711 similar to GP|17827234|dbj ORF1 {TT virus}, partial (3%) 38 0.028
>TC216673 similar to UP|O23575 (O23575) G2484-1 protein (Fragment), partial
(17%)
Length = 1381
Score = 489 bits (1260), Expect = e-138
Identities = 262/370 (70%), Positives = 291/370 (77%), Gaps = 1/370 (0%)
Frame = +1
Query: 696 WIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKVGAN 755
WIQESW EGVIT K KKDETT TVH PASGET V+RAW+LRPSLIWKDG+W++ SKVGAN
Sbjct: 13 WIQESWWEGVITAKKKKDETTFTVHFPASGETLVVRAWHLRPSLIWKDGKWIESSKVGAN 192
Query: 756 DSSTHKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESANPDEMRSLNLTENEIVFNIG 815
DSSTH+GDTP EKRPKLGS+AV+VKGKDKMSK DA ESA PDEM+ LNL EN+ VFNIG
Sbjct: 193 DSSTHEGDTPIEKRPKLGSHAVDVKGKDKMSKGSDAVESAKPDEMKLLNLAENDKVFNIG 372
Query: 816 KSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVNDKNDS 875
KSS NE+K D R VR+GLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAH +SK+ D+NDS
Sbjct: 373 KSSKNENKFDAHRMVRTGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHENSKIGDRNDS 552
Query: 876 VKIANFSMPQGSELRGWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRVNPPRNTSVSNT 935
VK+ NF MP S RGW+NSSKND+KEK GADSKPKT S+T
Sbjct: 553 VKLTNFLMPPSSGPRGWKNSSKNDAKEKHGADSKPKT--------------------SHT 672
Query: 936 EMNKDSSNHTKN-ASQSESRVERAPYSTTDGATQVPIVFSSQATSTNTLPTKRTFTSRAS 994
E KDSSN KN AS+SES+VERAP+S +DGAT P +FSS ATS + PTKR +SRAS
Sbjct: 673 ERIKDSSNLFKNAASKSESKVERAPHSASDGATG-PFLFSSLATSVDAHPTKRASSSRAS 849
Query: 995 KGKLAPASDKLRKGGGGKALNDKPTTSTSEPDALEPRRSNRRIQPTSRLLEGLQSSLMVS 1054
KGKLAPA K K KALND P S S D +EPRRSNRRIQPTSRLLEGLQSSL++S
Sbjct: 850 KGKLAPARVKSGKVEMEKALNDNPMKSAS--DMVEPRRSNRRIQPTSRLLEGLQSSLIIS 1023
Query: 1055 KIPSVSHNRN 1064
KIPSVSHNRN
Sbjct: 1024 KIPSVSHNRN 1053
>BE661568 similar to GP|20466528|gb G2484-1 protein {Arabidopsis thaliana},
partial (3%)
Length = 762
Score = 298 bits (764), Expect = 7e-81
Identities = 167/248 (67%), Positives = 188/248 (75%), Gaps = 1/248 (0%)
Frame = +2
Query: 196 GLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRAS 255
GLQS F GT S LDANNVTV PAQ +S PK KKRKK V+SED GQ+ LQSL P V S S
Sbjct: 2 GLQSIFTGTASLLDANNVTVSPAQHNSDPKPKKRKKVVVSEDLGQRALQSLAPGVGSHTS 181
Query: 256 TSVSAATPVGNVPMSSVEKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEA 315
T V+ PVGNVP++++EKSV+SVSPLADQ KND+ VEKRI+SDESLMKVKEARVHAEEA
Sbjct: 182 TPVAVVAPVGNVPITTIEKSVLSVSPLADQSKNDRNVEKRIMSDESLMKVKEARVHAEEA 361
Query: 316 SALSAAAVNHSLELWNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASN 375
SALSAAAVNHSLELWNQLDKHKNSG M DIEAKLASAAVA+AAAA +AKAAAAAANVASN
Sbjct: 362 SALSAAAVNHSLELWNQLDKHKNSGLMPDIEAKLASAAVAVAAAATIAKAAAAAANVASN 541
Query: 376 AAFQAKLMADEALISSGYENTSQGNNTFLPEGTSNLGQATPA-SILKGANGPNSPGSFIV 434
AA QAKLMADEA ++ G +N G++ P G NSPG +V
Sbjct: 542 AALQAKLMADEACFHLVMIIPAKVIKFLFLXGPNNFGKSYPCFPS*XXLMGINSPGLSMV 721
Query: 435 AAKEAIRR 442
AKE ++R
Sbjct: 722 LAKELVKR 745
>TC223850 similar to UP|Q8LPI8 (Q8LPI8) G2484-1 protein (At4g17330), partial
(3%)
Length = 510
Score = 224 bits (572), Expect = 1e-58
Identities = 116/160 (72%), Positives = 130/160 (80%), Gaps = 2/160 (1%)
Frame = +2
Query: 32 PQKHLCSQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPT 91
P+ L S+ +A RTSD KQS Q KGISSPLGR SSKATP I NPLIPLSSPLWSL T
Sbjct: 23 PETPLQSRSVA-RTSDLPHKQSAAQAKGISSPLGRTSSKATPPIVNPLIPLSSPLWSLST 199
Query: 92 LS--ADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSP 149
L +DSLQSSA+ARGSV+DY QA+TPLHPYQ+ RNFLGH+T W+SQ PLRGPWI SP
Sbjct: 200 LGLGSDSLQSSAIARGSVMDYPQAITPLHPYQTTPVRNFLGHNTPWMSQTPLRGPWIASP 379
Query: 150 TPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPG 189
TPA DN+T +SASP+SDTIKL SVKGSLPPSS IK+VT G
Sbjct: 380 TPATDNSTQISASPASDTIKLGSVKGSLPPSSGIKNVTSG 499
Score = 30.0 bits (66), Expect = 5.8
Identities = 12/16 (75%), Positives = 14/16 (87%)
Frame = +3
Query: 26 KNLILIPQKHLCSQDL 41
KNLIL+ QKHLC+ DL
Sbjct: 3 KNLILLTQKHLCNHDL 50
>CA801851 weakly similar to GP|20466528|gb G2484-1 protein {Arabidopsis
thaliana}, partial (8%)
Length = 445
Score = 205 bits (521), Expect = 1e-52
Identities = 110/149 (73%), Positives = 122/149 (81%), Gaps = 4/149 (2%)
Frame = +1
Query: 550 NEKNTRGQQAR-TVSDLVKPVDMVLGSESETQDPSFTVRNGSENLEENTF-KEGSLVEVF 607
NEKN+RG + R VS+LVKP+ +V GSE E Q P FTV NGSENL E++ KEG LVEVF
Sbjct: 1 NEKNSRGPKGRKVVSNLVKPIHVVPGSEPEIQAP-FTVNNGSENLVESSIIKEGLLVEVF 177
Query: 608 KDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVEG--PLKEWVSLECEGDKPPRIRTARP 665
KDEEG KAAWF NIL+L+D K YV YTSLVA EG PLKEWVSL C+GDK PRIRTARP
Sbjct: 178 KDEEGFKAAWFSANILTLRDDKAYVGYTSLVAAEGAGPLKEWVSLVCDGDKHPRIRTARP 357
Query: 666 LTSLQHEGTRKRRRAAMGDYAWSVGDRVD 694
L +LQ+EGTRKRRRAAMGDYAWSVGDRVD
Sbjct: 358 LNTLQYEGTRKRRRAAMGDYAWSVGDRVD 444
>BQ612937 similar to GP|20466528|gb G2484-1 protein {Arabidopsis thaliana},
partial (1%)
Length = 456
Score = 198 bits (504), Expect = 9e-51
Identities = 96/150 (64%), Positives = 117/150 (78%), Gaps = 2/150 (1%)
Frame = +2
Query: 615 AAWFMGNILSLKDGKVYVCYTSLVAVEG--PLKEWVSLECEGDKPPRIRTARPLTSLQHE 672
AAW+ +IL+LKDGK YVCY L+ EG PLKEW+SLE K PRIRT L L +E
Sbjct: 8 AAWYTASILNLKDGKAYVCYVVLLDDEGAGPLKEWISLEGGEVKSPRIRTPHYLPGLHNE 187
Query: 673 GTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRA 732
GTRKR+RAAM DY WSVGDRVDA +ESW+EGVIT++NKKD+ TLTVH P SG+T ++RA
Sbjct: 188 GTRKRQRAAMVDYTWSVGDRVDACSEESWQEGVITDQNKKDK-TLTVHFPVSGKTKLVRA 364
Query: 733 WNLRPSLIWKDGQWLDFSKVGANDSSTHKG 762
W+LRPS WKDG+W+++ KVG DSSTH+G
Sbjct: 365 WHLRPSRFWKDGKWIEYPKVGTGDSSTHEG 454
>TC232419
Length = 422
Score = 192 bits (489), Expect = 5e-49
Identities = 101/140 (72%), Positives = 111/140 (79%), Gaps = 3/140 (2%)
Frame = +1
Query: 529 ETSHAQNRDILSSEISASIMINEKNTRGQQA-RTVSDLVKPVDMVLGSESETQDPSFTVR 587
ETS+ NRDILS ISA I INEKN+RG + + VSDLVKP+D+V GSE E Q P FTV
Sbjct: 1 ETSYTHNRDILSGGISAPIKINEKNSRGAKGHKVVSDLVKPIDVVPGSEPEIQAPPFTVS 180
Query: 588 NGSENLEENTFKEGSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVE--GPLK 645
NGSENL E++ KEG LVEVFKDEEG KAAWF NIL+LKD K YV YTSLVA E GPLK
Sbjct: 181 NGSENLVESSIKEGLLVEVFKDEEGFKAAWFSANILTLKDNKAYVGYTSLVAAEGAGPLK 360
Query: 646 EWVSLECEGDKPPRIRTARP 665
EWVSLEC+GDKPPRIR ARP
Sbjct: 361 EWVSLECDGDKPPRIRAARP 420
>TC209716 similar to UP|O23575 (O23575) G2484-1 protein (Fragment), partial
(4%)
Length = 845
Score = 188 bits (478), Expect = 1e-47
Identities = 109/184 (59%), Positives = 129/184 (69%), Gaps = 10/184 (5%)
Frame = +2
Query: 890 RGWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRVNPPRNTSVSNTEMN---------KD 940
RGW+NSSKND+KEKLGADS+P K GK VLGRV PP+ +SN+ N KD
Sbjct: 5 RGWKNSSKNDTKEKLGADSRPTFKSGKSQSVLGRVVPPKENPLSNSRTNDLTSHAERIKD 184
Query: 941 SSNHTKNASQSESRVERAPYSTTDGATQVPIVFSSQATSTNTLPTKRTFTSRASKGKLAP 1000
SS+H KN SQSE++VERA YS + GA PI+ SS +ST++ P K+T TSRASKGKLAP
Sbjct: 185 SSSHFKNVSQSENQVERALYSGSTGAGAGPILHSSLVSSTDSHPAKKTSTSRASKGKLAP 364
Query: 1001 A-SDKLRKGGGGKALNDKPTTSTSEPDALEPRRSNRRIQPTSRLLEGLQSSLMVSKIPSV 1059
A +L K KA + P STSE EPRRS RRIQPTSRLLEGLQSSL++SKIPS
Sbjct: 365 AGGGRLGKIDEEKAFSGNPLKSTSE--NTEPRRSIRRIQPTSRLLEGLQSSLIISKIPSA 538
Query: 1060 SHNR 1063
SH +
Sbjct: 539 SHEK 550
>BE822011 similar to GP|20466528|gb G2484-1 protein {Arabidopsis thaliana},
partial (1%)
Length = 411
Score = 179 bits (454), Expect = 6e-45
Identities = 94/137 (68%), Positives = 108/137 (78%), Gaps = 2/137 (1%)
Frame = -1
Query: 75 IANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTS 134
IANPL+PLSSPLWSLPT S DSLQ SA ARGSVVDYSQALT HPYQ+P RNFLGH+TS
Sbjct: 405 IANPLMPLSSPLWSLPTPSCDSLQFSAFARGSVVDYSQALTXSHPYQTPPLRNFLGHNTS 226
Query: 135 WISQAPLRGPWIGSPTPAPDNN-THLSASPSSDTIKLASVKG-SLPPSSSIKDVTPGPPA 192
W+SQA L G W +PT APDNN +HL ASP +DTI+L+SVKG +PPSS IK+ PG PA
Sbjct: 225 WLSQATLCGAW--TPTSAPDNNSSHLXASPLTDTIRLSSVKGYPVPPSSGIKNAPPGLPA 52
Query: 193 SSSGLQSTFVGTDSQLD 209
S+GLQ+ F+ T LD
Sbjct: 51 XSTGLQNVFIATAPPLD 1
>BM140087 similar to GP|20466528|gb G2484-1 protein {Arabidopsis thaliana},
partial (6%)
Length = 538
Score = 164 bits (416), Expect = 2e-40
Identities = 82/100 (82%), Positives = 95/100 (95%)
Frame = +2
Query: 413 QATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENMDAILKAAELAAEAVS 472
+ATPASILKGANG NSPGS IV AKEA++RRVEAASAATKRAENMDAI+KAAELAAEAVS
Sbjct: 17 KATPASILKGANGTNSPGSIIVTAKEAVKRRVEAASAATKRAENMDAIVKAAELAAEAVS 196
Query: 473 QAGKIVTMGDPLPLIELIEAGPEGCWKASRESSREVGLLK 512
QAGKIVTMGDPLP+ +L+EAGPEGC KA+RESS++VG+++
Sbjct: 197 QAGKIVTMGDPLPISQLVEAGPEGCLKATRESSQQVGIIQ 316
>AW184980
Length = 447
Score = 78.2 bits (191), Expect = 2e-14
Identities = 38/49 (77%), Positives = 41/49 (83%)
Frame = +3
Query: 1023 SEPDALEPRRSNRRIQPTSRLLEGLQSSLMVSKIPSVSHNRNIPKGEHP 1071
S D +EPRRSNRRIQPTSRLLEGLQSSL++SKIPSVSHNRN HP
Sbjct: 6 SASDVVEPRRSNRRIQPTSRLLEGLQSSLIISKIPSVSHNRNTKSEYHP 152
>TC207092 similar to UP|Q94ET8 (Q94ET8) LCI5, partial (11%)
Length = 1015
Score = 41.6 bits (96), Expect = 0.002
Identities = 45/149 (30%), Positives = 64/149 (42%), Gaps = 26/149 (17%)
Frame = +1
Query: 72 TPTIANPLIPL-------------------SSPLWSLPTLSADSLQSSALARGSVVDYSQ 112
TPT+ LIP+ S P S+PT A + SS R S S
Sbjct: 61 TPTLTLTLIPILILITTLTLILTPKTSSFCSPPTASVPTAVA-AAASSTPPRSSSSSSSP 237
Query: 113 ALTPLHPYQSPSPRNFLGHSTSWISQA-PLRGPWIGSPTPAPDNNTHLSASPS------S 165
A PL P+ R+ L S S S + P RGP+ SP+P+P +T ++SPS S
Sbjct: 238 A--PLSSSTPPTRRSALPESDSTASGSEPTRGPFSTSPSPSPLKSTIETSSPSPMTPSPS 411
Query: 166 DTIKLASVKGSLPPSSSIKDVTPGPPASS 194
+ A+ S P +++ PP S+
Sbjct: 412 PSATAAASSASSPAAAAAASGRVAPPTST 498
Score = 36.2 bits (82), Expect = 0.081
Identities = 37/135 (27%), Positives = 53/135 (38%), Gaps = 3/135 (2%)
Frame = +1
Query: 153 PDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSS--GLQSTFVGTDSQLDA 210
P ++ S +S +A+ S PP SS +P P +SS+ +S +DS
Sbjct: 130 PKTSSFCSPPTASVPTAVAAAASSTPPRSSSSSSSPAPLSSSTPPTRRSALPESDSTASG 309
Query: 211 NNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNV-PM 269
+ T P S P K + S A AS AS+ +AA G V P
Sbjct: 310 SEPTRGPFSTSPSPSPLKSTIETSSPSPMTPSPSPSATAAASSASSPAAAAAASGRVAPP 489
Query: 270 SSVEKSVVSVSPLAD 284
+S + SPL D
Sbjct: 490 TSTPR-----SPLTD 519
>TC216940 weakly similar to GB|AAP37755.1|30725466|BT008396 At1g68580
{Arabidopsis thaliana;} , partial (11%)
Length = 1049
Score = 40.8 bits (94), Expect = 0.003
Identities = 19/55 (34%), Positives = 28/55 (50%)
Frame = +2
Query: 693 VDAWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWL 747
VDAW + W EG++ +K+ E+ V+ P SV NLR S W +W+
Sbjct: 11 VDAWWHDGWWEGIVVQKD--SESNCHVYFPGEKVVSVFGPGNLRESQDWVGNEWV 169
>BF071113
Length = 213
Score = 40.8 bits (94), Expect = 0.003
Identities = 19/33 (57%), Positives = 23/33 (69%)
Frame = +2
Query: 9 GTCGRMSGVSAWKGSVVKNLILIPQKHLCSQDL 41
G GRM G+ AWK S+VK+LIL K LC+ DL
Sbjct: 95 GAFGRMPGLPAWKSSMVKSLIL*TWKRLCNHDL 193
>TC226650 similar to UP|Q94BZ2 (Q94BZ2) At1g30970/F17F8_14, partial (65%)
Length = 1596
Score = 39.7 bits (91), Expect = 0.007
Identities = 45/190 (23%), Positives = 78/190 (40%), Gaps = 6/190 (3%)
Frame = +1
Query: 40 DLAARTSDSTVKQSVLQGKGISSPLG-----RASSKATPTIANPLIPLSSPLWSLPTLSA 94
D+ ++ + + + L G I PLG R + P + NP +P+ W +P
Sbjct: 394 DVPSKAAKVDIPPTQLVGGMIPPPLGTGYPPRPTLPTMPPMYNPAVPVPPNAWMVPPRPQ 573
Query: 95 DSL-QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAP 153
Q A++ Y+Q PL P Q+ P +T+ Q + P + + P P
Sbjct: 574 PWFSQPPAVSVPPAAPYTQQ--PLFPVQNVRPPL---PATAPALQTQITPPGLPTSAPVP 738
Query: 154 DNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNV 213
+ ++ T +S + P SS+ VTP + ST V D+ L +N+
Sbjct: 739 VSQPLFPVVGNNHTTTQSSAFSAPPLPSSVPSVTP--------VMSTNVPVDTHLSSNS- 891
Query: 214 TVPPAQQSSG 223
+V + Q+ G
Sbjct: 892 SVTSSYQAIG 921
>TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, partial (8%)
Length = 1061
Score = 39.7 bits (91), Expect = 0.007
Identities = 57/237 (24%), Positives = 84/237 (35%), Gaps = 29/237 (12%)
Frame = +1
Query: 81 PLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGH---STSWIS 137
P S P++ S L +S S S + T +P +P +F ST W S
Sbjct: 112 PKSCPVYR*SR*SGCGLPASGGGPSSTAGTSSSSTSTNP----TPASFSATAPTSTPWTS 279
Query: 138 QAPLRGPW---------------IGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSS 182
R PW PT + + T L+ SPS T+ AS S P +S+
Sbjct: 280 NLRNRIPWN*ATP*CATATASRSSAPPTASSASPTLLTTSPSG-TLSSASTVFSPPTAST 456
Query: 183 IKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSS-----------GPKAKKRKK 231
+ PPAS+ A+ T PP SS P +
Sbjct: 457 APNPPSSPPAST---------------ASATTPPPTTTSS*ALPTSWISRNAPSIPRSSS 591
Query: 232 DVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSPLADQPKN 288
+ G+ TP+ A S S A G+ P +S + +S SPL + ++
Sbjct: 592 TPSNPTPGRTSPACPTPSAAPAPWASSSPAPSTGSSPENSNPTNPISSSPLTSRARH 762
>TC213615 similar to UP|Q9SUL7 (Q9SUL7) SNF1 like protein kinase
(CBL-interacting protein kinase 4), partial (45%)
Length = 674
Score = 38.9 bits (89), Expect = 0.012
Identities = 46/167 (27%), Positives = 70/167 (41%), Gaps = 3/167 (1%)
Frame = +3
Query: 32 PQKHLCSQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSL-- 89
P + S AA T+ T +S S P R++S +T +A P SSP +
Sbjct: 225 PASCVRSTPCAASTTTPTSSKSTRS----SPPRPRSTSSSTSPVAASSSP-SSPAAAASR 389
Query: 90 -PTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGS 148
P+ +A S SS L+ + S T H S +P A R P S
Sbjct: 390 SPSPAATSRSSSPLSASATAMASPTETSSHRTSSSTP------------PATSRSPTSAS 533
Query: 149 PTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSS 195
P P+ + +T +S++P + + S +++ TPGP ASSS
Sbjct: 534 P-PSRNTSTTVSSTPPAARRPSPRQRSSAASATTAXKPTPGPAASSS 671
>TC216015 similar to UP|Q9M610 (Q9M610) Ttg1-like protein, partial (91%)
Length = 1151
Score = 38.5 bits (88), Expect = 0.016
Identities = 60/219 (27%), Positives = 93/219 (42%), Gaps = 18/219 (8%)
Frame = +2
Query: 63 PLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQ- 121
P S + TP++ +PL LS+ S P S S+A ++ + + +L+ L P++
Sbjct: 41 PKNPISDRKTPSLTSPLT-LSTACHSPPPTPTAS-PSAASSKNTTTASTSSLSTLTPFR* 214
Query: 122 -----SPSPRNFLG-HSTSWISQAPLRGPWIGSPTPAPDNNTHLSA------SPSSDTIK 169
SPS L +S S + PL P SP PA + + S SPSS T +
Sbjct: 215 LPTLLSPSTTLTLPPNSCSTPANPPLPLPPTSSPPPATTSASGRSVITPWMPSPSSTTAR 394
Query: 170 LASVKGSLPPS---SSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKA 226
AS PPS +S +P PPAS+ S GT + P+ + + +
Sbjct: 395 PASSAPP*PPSTGTTSTPTASP-PPASTPPAPS---GTSN---------APSSKPNSSRT 535
Query: 227 KKRKKDVLSEDHGQKLLQS--LTPAVASRASTSVSAATP 263
KR + HG++ S P +ST + +TP
Sbjct: 536 TKRS----TTSHGERPESSPPSPPTAPLESSTFATRSTP 640
>TC228613 weakly similar to UP|PE31_ARATH (Q9LHA7) Peroxidase 31 precursor
(Atperox P31) (ATP41) , partial (50%)
Length = 536
Score = 38.1 bits (87), Expect = 0.021
Identities = 39/119 (32%), Positives = 58/119 (47%), Gaps = 3/119 (2%)
Frame = +3
Query: 60 ISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHP 119
+SSP S PT + P SSP + P+ S SSA A + LT P
Sbjct: 12 LSSPAPTPSP--APTSSPPPPATSSPCSAAPS----SPSSSAAATAAPPSPPPFLTTSPP 173
Query: 120 YQSPSPRNFLGHSTSWISQAPLRGPWIG-SPTPAPDNNTHLSASPSSDT--IKLASVKG 175
Q PSP++ H +S + + + W+ PTPA D+ T ++SP+S T LA+++G
Sbjct: 174 PQCPSPKS---HKSSPTAASQSKNSWLSVGPTPA-DSPTAPNSSPTSPTPPTTLATLRG 338
>TC207392 similar to UP|BNK_DROME (P40794) Bottleneck protein, partial (7%)
Length = 1544
Score = 38.1 bits (87), Expect = 0.021
Identities = 30/143 (20%), Positives = 60/143 (40%), Gaps = 4/143 (2%)
Frame = +2
Query: 888 ELRGWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRVNPPR----NTSVSNTEMNKDSSN 943
E + + N + L K +F +PP N + T++S+ K +
Sbjct: 119 ESKALSSGGSNSRRRMLLEFDAEKEEFEQPPKTKSTKNQTKLIKPTTNLSSKNQTKTIKS 298
Query: 944 HTKNASQSESRVERAPYSTTDGATQVPIVFSSQATSTNTLPTKRTFTSRASKGKLAPASD 1003
+ N+S+++ ++ + + D T V + + ++ TL +K+ + SKG +S
Sbjct: 299 NNLNSSKNQLKLAKTKAAAGDTLTNTTEVVAVKKLNSTTLKSKKL--NSTSKGLTKSSSL 472
Query: 1004 KLRKGGGGKALNDKPTTSTSEPD 1026
L K GGK K T + + +
Sbjct: 473 DLAKTSGGKNKTTKATATNKDKE 541
>BU964711 similar to GP|17827234|dbj ORF1 {TT virus}, partial (3%)
Length = 449
Score = 37.7 bits (86), Expect = 0.028
Identities = 31/110 (28%), Positives = 48/110 (43%), Gaps = 1/110 (0%)
Frame = +3
Query: 122 SPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSS 181
SP P ++ +TS S+ P SPTP P ++ S++ +S + AS PSS
Sbjct: 144 SPQPPHYKAAATSSNSRTLFTPPTSPSPTPTPTSSPPASSTLASSSTSPASTPPPPSPSS 323
Query: 182 SIKDVTP-GPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRK 230
+ +P G PAS + L S +PP P+ ++RK
Sbjct: 324 AASTASPSGDPASGTPL--------SWYQVRGFALPPPP----PRRRRRK 437
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.308 0.125 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,845,346
Number of Sequences: 63676
Number of extensions: 614933
Number of successful extensions: 4368
Number of sequences better than 10.0: 194
Number of HSP's better than 10.0 without gapping: 3798
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4118
length of query: 1091
length of database: 12,639,632
effective HSP length: 107
effective length of query: 984
effective length of database: 5,826,300
effective search space: 5733079200
effective search space used: 5733079200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC137077.8


