

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136954.5 - phase: 0
(362 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
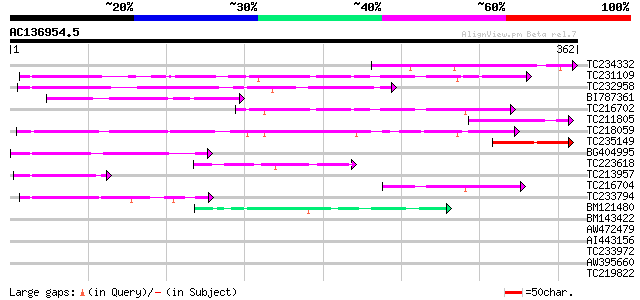
Score E
Sequences producing significant alignments: (bits) Value
TC234332 88 5e-18
TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, par... 75 4e-14
TC232958 75 5e-14
BI787361 69 3e-12
TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like pro... 65 4e-11
TC211805 similar to UP|Q9K6E7 (Q9K6E7) BH3782 protein, partial (... 60 2e-09
TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%) 55 5e-08
TC235149 46 3e-05
BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa ... 45 4e-05
TC223618 45 7e-05
TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 44 1e-04
TC216704 similar to UP|Q7PIN1 (Q7PIN1) ENSANGP00000023661 (Fragm... 43 2e-04
TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 43 3e-04
BM121480 41 7e-04
BM143422 41 0.001
AW472479 weakly similar to PIR|T10479|T104 glycine-rich RNA-bind... 37 0.014
AI443156 36 0.024
TC233972 weakly similar to GB|AAP37713.1|30725382|BT008354 At3g2... 36 0.031
AW395660 35 0.053
TC219822 homologue to UP|CENB_HUMAN (P07199) Major centromere au... 34 0.091
>TC234332
Length = 644
Score = 88.2 bits (217), Expect = 5e-18
Identities = 52/140 (37%), Positives = 75/140 (53%), Gaps = 9/140 (6%)
Frame = +2
Query: 232 DIKYDGVYLSNSISWLVCHRYKC--QQKNLTTEQFVIISLDLETETYTQLQLP----KLP 285
++ DG + +++WL Q + +T + VI S DL+ E+Y L +P ++P
Sbjct: 5 EVSQDGASVRGTVNWLALPNSSSDYQWETVTIDDLVIFSYDLKNESYRYLLMPDGLLEVP 184
Query: 286 FDNPNICALMDCICFSYDFKETHFVIWQMKEFGVEESWTQFLKISYQNLGINYILGRNGF 345
P + L C+C S+ HF W MKEFGVE+SWT+FL ISY L I+ GF
Sbjct: 185 HSPPELVVLKGCLCLSHRHGGNHFGFWLMKEFGVEKSWTRFLNISYDQLHIH-----GGF 349
Query: 346 LVLPV--CLSE-NGATLIGN 362
L PV C+SE +G L+ N
Sbjct: 350 LDHPVILCMSEDDGVVLLEN 409
>TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, partial (8%)
Length = 1061
Score = 75.5 bits (184), Expect = 4e-14
Identities = 91/337 (27%), Positives = 144/337 (42%), Gaps = 10/337 (2%)
Frame = +3
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNK 66
LPVE++T EIL R V S++ L+ K W ++I FV HL S S LR
Sbjct: 93 LPVEVVT-EILSRLPVKSVIRLRSTCKWWRSIIDSRHFVLFHLNKSHSSLILR------- 248
Query: 67 NLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCV-DCCGIIGSCNG 125
R Y SL L+S + P+ +L + C + ++GS NG
Sbjct: 249 ---------HRSHLY---------SLDLKSPEQN-PV--ELSHPLMCYSNSIKVLGSSNG 365
Query: 126 LICLHGCFHGSGYKKHSFCFWNPATRSKSKTL----LYVP-SYLNRVRL-GFGYDNSTDT 179
L+C+ WNP R K + L + P S L R+ GFG+ + ++
Sbjct: 366 LLCISNV-------ADDIALWNPFLR-KHRILPADRFHRPQSSLFAARVYGFGHHSPSND 521
Query: 180 YKTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWDDIKYDGVY 239
YK + +D R + V+++TL W+++ S P L GV+
Sbjct: 522 YKLLSITYFVDLQ---KRTFDSQVQLYTLKSDSWKNLPS-MPYALCCARTM------GVF 671
Query: 240 LSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLPKL---PFDNPNICALMD 296
+S S+ WLV + + + +L I+S DL ET+ ++ LP FD + L
Sbjct: 672 VSGSLHWLVTRKLQPHEPDL------IVSFDLTRETFHEVPLPVTVNGDFDM-QVALLGG 830
Query: 297 CICFSYDFKETHFVIWQMKEFGVEESWTQFLKISYQN 333
C+C + + T F +W M+ +G SW + + N
Sbjct: 831 CLCV-VEHRGTGFDVWVMRVYGSRNSWEKLFTLLENN 938
>TC232958
Length = 758
Score = 75.1 bits (183), Expect = 5e-14
Identities = 71/250 (28%), Positives = 103/250 (40%), Gaps = 8/250 (3%)
Frame = +1
Query: 6 ILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSN 65
+LP +LIT EILLR V SL+ K V K W LISDP F K H L + + L + S+
Sbjct: 58 VLPQDLIT-EILLRLPVKSLVRFKSVCKSWLFLISDPRFAKSHFDLFAALADRILFIASS 234
Query: 66 KNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGSCNG 125
+ + L ++S+ + DL IIGSC G
Sbjct: 235 ----------------APELRSIDFNASLHDDSASVAVTVDLPAPKPYFHFVEIIGSCRG 366
Query: 126 LICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVPSYLNR------VRLGFGYDNSTDT 179
I LH H C WNP T K + P + N+ + GFGYD STD
Sbjct: 367 FILLHCLSH--------LCVWNPTT-GVHKVVPLSPIFFNKDAVFFTLLCGFGYDPSTDD 519
Query: 180 YKTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQS-SFP-VELALRSRWDDIKYDG 237
+ V + + ++F+L + W+ I+ FP +R++ G
Sbjct: 520 FLVV-------HACYNPKHQANCAEIFSLRANAWKGIEGIHFPYTHFRYTNRYNQF---G 669
Query: 238 VYLSNSISWL 247
+L+ +I WL
Sbjct: 670 SFLNGAIHWL 699
>BI787361
Length = 423
Score = 68.9 bits (167), Expect = 3e-12
Identities = 46/129 (35%), Positives = 69/129 (52%), Gaps = 2/129 (1%)
Frame = +3
Query: 24 SLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNKNLRLQIRA-GGRGCSYT 82
+LM ++VS+ WN+LI DP FVK+HL+ S N ++ L+ +A R
Sbjct: 3 ALMRFRYVSETWNSLIFDPTFVKLHLERSP----------KNTHVLLEFQAIYDRDVGQQ 152
Query: 83 VTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGSCNGLICLHGCFHGSGYKKH- 141
V VAP S+ L+E+ + +I DD F + I GSCNGL+C+ CF +++
Sbjct: 153 VGVAPCSIRRLVENPSFTI---DDCLTLFKHTN--SIFGSCNGLVCMTKCFDVREFEEEC 317
Query: 142 SFCFWNPAT 150
+ WNPAT
Sbjct: 318 QYRLWNPAT 344
>TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like protein,
partial (5%)
Length = 1631
Score = 65.5 bits (158), Expect = 4e-11
Identities = 53/192 (27%), Positives = 90/192 (46%), Gaps = 13/192 (6%)
Frame = +3
Query: 145 FWNPATRSKSKTLLYVP--------SYLNRVRL-GFGYDNSTDTYKTVMFGITMDEGLGG 195
FWNP+ R + + L Y+P + L R+ GFG+D+ T YK V +D
Sbjct: 36 FWNPSLR-QHRILPYLPVPRRRHPDTTLFAARVCGFGFDHKTRDYKLVRISYFVDLH--- 203
Query: 196 NRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWDDIKYDGVYLSNSISWLVCHRYKCQ 255
+R A VK++TL + W+ + S P L GV++ NS+ W+V + +
Sbjct: 204 DRSFDAQVKLYTLRANAWKTLPS-LPYALCCARTM------GVFVGNSLHWVVTRKLEPD 362
Query: 256 QKNLTTEQFVIISLDLETETYTQLQLPKLPFDNP----NICALMDCICFSYDFKETHFVI 311
Q +L II+ DL + + +L LP + ++ L +C + +F +T +
Sbjct: 363 QPDL------IIAFDLTHDIFRELPLPDTGGVDGGFEIDLALLGGSLCMTVNFHKTRIDV 524
Query: 312 WQMKEFGVEESW 323
W M+E+ +SW
Sbjct: 525 WVMREYNRRDSW 560
>TC211805 similar to UP|Q9K6E7 (Q9K6E7) BH3782 protein, partial (13%)
Length = 693
Score = 59.7 bits (143), Expect = 2e-09
Identities = 26/67 (38%), Positives = 38/67 (55%)
Frame = +1
Query: 294 LMDCICFSYDFKETHFVIWQMKEFGVEESWTQFLKISYQNLGINYILGRNGFLVLPVCLS 353
L C+C +YD+K+THFV+W MK++G ESW + + I Y N+ P +S
Sbjct: 13 LQGCLCMNYDYKKTHFVVWMMKDYGARESWVKLVSIPYVPNPENFSYSG------PYYIS 174
Query: 354 ENGATLI 360
ENG L+
Sbjct: 175 ENGEVLL 195
>TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%)
Length = 1185
Score = 55.1 bits (131), Expect = 5e-08
Identities = 75/341 (21%), Positives = 141/341 (40%), Gaps = 20/341 (5%)
Frame = +3
Query: 5 EILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFS 64
E LP EL++ +L R L+L K V K W LI+DP FV + + S +
Sbjct: 60 EHLPGELVSN-VLSRLPSKVLLLCKCVCKSWFDLITDPHFVSNYYVVYNSLQS------Q 218
Query: 65 NKNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGSCN 124
++L + R G ++V + + + +S + + +Y I+G CN
Sbjct: 219 EEHLLVIRRPFFSGLKTYISVLSWNTNDPKKHVSSDV-LNPPYEYNSDHKYWTEILGPCN 395
Query: 125 GLICLHGCFHGSGYKKHSFCFWNPAT---RSKSKTLLYVP--SYLNRVRLGFGYDNSTDT 179
G+ L G + NP+ ++ K+ P +Y GFG+D T+
Sbjct: 396 GIYFLEG---------NPNVLMNPSLGEFKALPKSHFTSPHGTYTFTDYAGFGFDPKTND 548
Query: 180 YKTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSF---PVELALRSRWDDIKYD 236
YK V+ + + ++++L + WR + S P+E+ SR
Sbjct: 549 YKVVVLKDLWLKETDEREIGYWSAELYSLNSNSWRKLDPSLLPLPIEIWGSSRVF----- 713
Query: 237 GVYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLPKL------------ 284
Y +N CH + +++ T+ V+++ D+ E++ ++++PK+
Sbjct: 714 -TYANNC-----CHWWGFVEESDATQD-VVLAFDMVKESFRKIRVPKIRDSSDEKFGTLV 872
Query: 285 PFDNPNICALMDCICFSYDFKETHFVIWQMKEFGVEESWTQ 325
PF+ A + + + E F +W MK++ E SW +
Sbjct: 873 PFEES---ASIGFLVYPVRGTEKRFDVWVMKDYWDEGSWVK 986
>TC235149
Length = 435
Score = 45.8 bits (107), Expect = 3e-05
Identities = 22/52 (42%), Positives = 32/52 (61%)
Frame = +2
Query: 309 FVIWQMKEFGVEESWTQFLKISYQNLGINYILGRNGFLVLPVCLSENGATLI 360
FV+W +EFGVE SWT+ L +SY++ N+ V P+C+SEN L+
Sbjct: 2 FVVWLTREFGVERSWTRLLNVSYEHFR-NHGCPPYYRFVTPLCMSENEDVLL 154
>BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 394
Score = 45.4 bits (106), Expect = 4e-05
Identities = 38/129 (29%), Positives = 61/129 (46%)
Frame = +1
Query: 1 MNISEILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRL 60
+ +S+ LP E++T EIL R V SL+ + SK W +LI +HL S L
Sbjct: 22 LTMSDHLPREVLT-EILSRLPVRSLLRFRSTSKSWKSLIDSQHLNWLHLTRS-------L 177
Query: 61 ALFSNKNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGII 120
L SN +L L++ + ++ P S++ L ++SI ++
Sbjct: 178 TLASNTSLILRVDSDLYQTNFPTLDPPVSLNHPLMCYSNSIT----------------LL 309
Query: 121 GSCNGLICL 129
GSCNGL+C+
Sbjct: 310 GSCNGLLCI 336
>TC223618
Length = 444
Score = 44.7 bits (104), Expect = 7e-05
Identities = 35/110 (31%), Positives = 45/110 (40%), Gaps = 6/110 (5%)
Frame = +1
Query: 118 GIIGSCNGLICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVPSYLNRVR------LGF 171
GI+GSCNGL+C K WNP+ R K+ P N R G
Sbjct: 124 GIVGSCNGLLCF-------AIKGDCVLLWNPSIRVSKKS----PPLGNNWRPGCFTAFGL 270
Query: 172 GYDNSTDTYKTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFP 221
GYD+ + YK V E VKV+++ + WR IQ FP
Sbjct: 271 GYDHVNEDYKVVAVFCDPSE-----YFIECKVKVYSMATNSWRKIQ-DFP 402
>TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 696
Score = 43.9 bits (102), Expect = 1e-04
Identities = 30/63 (47%), Positives = 36/63 (56%)
Frame = +3
Query: 3 ISEILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLAL 62
+S LP+ELI EILLR V S++ K V K W +LISDP F L S + RL L
Sbjct: 21 LSVTLPLELIR-EILLRSPVRSVLRFKCVCKSWLSLISDPQFTHFDLAASPTH---RLIL 188
Query: 63 FSN 65
SN
Sbjct: 189ISN 197
>TC216704 similar to UP|Q7PIN1 (Q7PIN1) ENSANGP00000023661 (Fragment),
partial (5%)
Length = 1232
Score = 43.1 bits (100), Expect = 2e-04
Identities = 23/95 (24%), Positives = 46/95 (48%), Gaps = 4/95 (4%)
Frame = +2
Query: 239 YLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLPKLPFDNP----NICAL 294
++ NS+ W+V + + Q +L I++ DL E +T+L LP ++ L
Sbjct: 2 FVGNSLHWVVTRKLEPDQPDL------IVAFDLTHEIFTELPLPDTGGVGGGFEIDVALL 163
Query: 295 MDCICFSYDFKETHFVIWQMKEFGVEESWTQFLKI 329
D +C + +F + +W M+E+ +SW + +
Sbjct: 164 GDSLCMTVNFHNSKMDVWVMREYNRGDSWCKLFTL 268
>TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 449
Score = 42.7 bits (99), Expect = 3e-04
Identities = 47/128 (36%), Positives = 59/128 (45%), Gaps = 4/128 (3%)
Frame = +1
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNK 66
LP ELI EILLR V SL+ K V K + +LISDP FV H L+ S + RL L S+
Sbjct: 109 LPQELIR-EILLRLPVKSLLRFKCVCKSFLSLISDPQFVISHYALAASPTH-RLILRSHD 282
Query: 67 NLRLQIRAGG--RGCSYTVTVAPTSVSLLLESTTSSIPI--ADDLQYQFSCVDCCGIIGS 122
I + CS V P + SIP DD + I+GS
Sbjct: 283 FYAQSIATESVFKTCSRDVVYFPLPL--------PSIPCLRLDDFGIRPK------ILGS 420
Query: 123 CNGLICLH 130
C GL+ L+
Sbjct: 421 CRGLVLLY 444
>BM121480
Length = 917
Score = 41.2 bits (95), Expect = 7e-04
Identities = 44/167 (26%), Positives = 65/167 (38%), Gaps = 3/167 (1%)
Frame = +3
Query: 119 IIGSCNGLICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVPSYLNRVRLGFGYDNSTD 178
I+GSC GL+ L+ + S + WNP+ + K L + GFGYD S D
Sbjct: 234 ILGSCRGLVLLY--YDDSA----NLILWNPSL-GRHKXLPNYRDDITSFPYGFGYDESKD 392
Query: 179 TYKTVMFGITM---DEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWDDIKY 235
Y ++ G+ + G +T K D+I D + E D I
Sbjct: 393 EYLLILIGLPKFGPETGADIYSFKTESWKT----DTIVYDPLXRYXAE-------DXIAR 539
Query: 236 DGVYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLP 282
G L+ ++ W V K VII+ DL T + + LP
Sbjct: 540 AGSLLNGALHWFVFSESK--------XDHVIIAFDLVERTLSXIPLP 656
>BM143422
Length = 424
Score = 40.8 bits (94), Expect = 0.001
Identities = 27/65 (41%), Positives = 38/65 (57%)
Frame = +1
Query: 2 NISEILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLA 61
N ++ LP++LI ILL+ V S+ K V K W +LISDP F H L+ + + RL
Sbjct: 214 NHTQTLPLDLIEL-ILLKLPVKSVTRFKCVCKSWLSLISDPQFGFSHFDLALAVPSHRLL 390
Query: 62 LFSNK 66
L SN+
Sbjct: 391 LRSNE 405
>AW472479 weakly similar to PIR|T10479|T104 glycine-rich RNA-binding protein
GRP1 - Commerson's wild potato, partial (22%)
Length = 414
Score = 37.0 bits (84), Expect = 0.014
Identities = 33/134 (24%), Positives = 55/134 (40%), Gaps = 2/134 (1%)
Frame = +2
Query: 91 SLLLESTTSSIPIADDLQYQFSCVDCCGIIGSCNGLICLHGCFHGSGYKKHSFCFWNPAT 150
SL L T +P DL++ ++ CNGLIC+ G + NP+
Sbjct: 59 SLTLSYTLLRLPSFPDLEFP--------VLSFCNGLICI-----AYGERCLPIIICNPSI 199
Query: 151 RSK--SKTLLYVPSYLNRVRLGFGYDNSTDTYKTVMFGITMDEGLGGNRMRTAVVKVFTL 208
R T P Y N + G+D++ YK + +D+ G + +V++++L
Sbjct: 200 RRYVWLPTPHDYPGYYNSC-IALGFDSTNCDYKVIRISCIVDDESFG--LSAPLVELYSL 370
Query: 209 GDSIWRDIQSSFPV 222
WR + PV
Sbjct: 371 KSGSWRILDGIAPV 412
>AI443156
Length = 414
Score = 36.2 bits (82), Expect = 0.024
Identities = 38/143 (26%), Positives = 56/143 (38%), Gaps = 1/143 (0%)
Frame = +3
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNK 66
LPVE++T EIL R V S++ L+ K W ++I F+ HL +K + L L
Sbjct: 90 LPVEVVT-EILSRLPVKSVIRLRSTCKWWRSIIDSRHFILFHL----NKSHTSLILRHRS 254
Query: 67 NLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCV-DCCGIIGSCNG 125
L SL L+S P +L + C + ++GS NG
Sbjct: 255 QL---------------------YSLDLKSLLDPNPF--ELSHPLMCYSNSIKVLGSSNG 365
Query: 126 LICLHGCFHGSGYKKHSFCFWNP 148
L+C+ WNP
Sbjct: 366 LLCISNV-------XDDIALWNP 413
>TC233972 weakly similar to GB|AAP37713.1|30725382|BT008354 At3g23880
{Arabidopsis thaliana;} , partial (10%)
Length = 631
Score = 35.8 bits (81), Expect = 0.031
Identities = 23/44 (52%), Positives = 27/44 (61%)
Frame = -2
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLK 50
LP E+I EILLR V SL KFV K +LIS+P F K H +
Sbjct: 540 LPQEIII-EILLRLPVKSLPSFKFVCKS*LSLISNPHFAKWHFE 412
>AW395660
Length = 381
Score = 35.0 bits (79), Expect = 0.053
Identities = 20/40 (50%), Positives = 25/40 (62%)
Frame = +3
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVK 46
LP EL+ EIL R V SL+ + V K W +LI DP F+K
Sbjct: 264 LPDELVV-EILSRLPVKSLLQFRCVCKSWMSLIYDPYFMK 380
>TC219822 homologue to UP|CENB_HUMAN (P07199) Major centromere autoantigen B
(Centromere protein B) (CENP-B), partial (5%)
Length = 710
Score = 34.3 bits (77), Expect = 0.091
Identities = 19/45 (42%), Positives = 27/45 (59%)
Frame = +1
Query: 10 ELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKS 54
E + EIL+ V++ + L+ V K W +L+ DP FVK HL S S
Sbjct: 202 EGLMIEILVWIRVSNPLQLRCVCKRWKSLVVDPQFVKKHLHTSLS 336
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.140 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,655,381
Number of Sequences: 63676
Number of extensions: 336602
Number of successful extensions: 1829
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 1806
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1818
length of query: 362
length of database: 12,639,632
effective HSP length: 98
effective length of query: 264
effective length of database: 6,399,384
effective search space: 1689437376
effective search space used: 1689437376
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC136954.5


