

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136953.9 - phase: 0
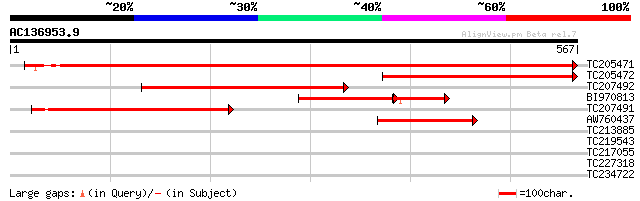
(567 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC205471 similar to UP|Q8GZD8 (Q8GZD8) Neutral leucine aminopept... 827 0.0
TC205472 similar to UP|Q43034 (Q43034) Leucine aminopeptidase (C... 362 e-100
TC207492 similar to UP|Q944P7 (Q944P7) AT4g30920/F6I18_170, part... 325 4e-89
BI970813 similar to PIR|T14912|T14 leucyl aminopeptidase (EC 3.4... 172 1e-65
TC207491 similar to UP|O65557 (O65557) Leucyl aminopeptidase-lik... 238 5e-63
AW760437 similar to GP|27463709|gb| neutral leucine aminopeptida... 142 4e-34
TC213885 weakly similar to UP|Q9VUB7 (Q9VUB7) CG32133-PA, partia... 30 3.8
TC219543 weakly similar to UP|Q94CH8 (Q94CH8) Family II lipase E... 30 3.8
TC217055 similar to UP|O64418 (O64418) Aldehyde oxidase , partia... 29 4.9
TC227318 similar to UP|Q75H88 (Q75H88) Expressed protein, 3'-par... 29 6.4
TC234722 similar to UP|Q6WAY9 (Q6WAY9) Pol (Fragment), partial (... 28 8.4
>TC205471 similar to UP|Q8GZD8 (Q8GZD8) Neutral leucine aminopeptidase
preprotein precursor , partial (87%)
Length = 1997
Score = 827 bits (2137), Expect = 0.0
Identities = 427/564 (75%), Positives = 481/564 (84%), Gaps = 11/564 (1%)
Frame = +2
Query: 15 SSSSSLFLTS----------RIRFASFPKRSFHSTTKLMSQSRYTTLGLTHPTNIEAPKI 64
SS SSLFL + R+ FAS P R +LM+++ TLGLT P N E+ K+
Sbjct: 65 SSFSSLFLKTLTLRSSTPFQRLSFASQPLR------RLMARA---TLGLTLPANTESLKV 217
Query: 65 SFSATDVDVTEWKGDILAVGVTEKDLTRDAKSRFENSILNKIDLKLDGLLSEASSEEDFS 124
SF+A ++DV EWKGD+LAVGVTEKD+ R+ +SRFEN+IL+ +D KL GLL + SSEEDFS
Sbjct: 218 SFAAKELDVAEWKGDLLAVGVTEKDVARNVESRFENAILSALDSKLGGLLGQVSSEEDFS 397
Query: 125 GKVGQSTVVRI-KGLGSKRVGLIGLGQLPSTTALYKGLGEAVVAVAKSAQASNVAIVLAS 183
GK+GQSTV+RI GLG KRV L+GLG STT+ +K L EA A AKSAQA +VA+ LAS
Sbjct: 398 GKLGQSTVLRIASGLGFKRVALLGLGASASTTSAFKSLREAGAAAAKSAQAGHVAVALAS 577
Query: 184 SEGLSSESKLSTAYAIASGAVLGLFEDQRYKSESKKPAVRSIDIIGLGTGPDLEKKLKYA 243
+ LS+ESK TA AIASG +LG FED RYKSESKK + S+DIIGLGTGP+LEKKLKYA
Sbjct: 578 PDALSTESKPKTASAIASGTLLGTFEDNRYKSESKKSVLSSVDIIGLGTGPELEKKLKYA 757
Query: 244 GDVSFGIIFGRELVNSPANVLTPGVLAEEASKVASTYSDVFTAKILDADQCKELKMGSYL 303
D+S GII GRELVNSPANVLTPGVLAE ASK+ASTY+DVFTA +L A+QCKELKMGSYL
Sbjct: 758 ADISSGIILGRELVNSPANVLTPGVLAEAASKIASTYNDVFTAILLYAEQCKELKMGSYL 937
Query: 304 GVAAASANPPRFIHLTYKPPSGSVKVKLALVGKGLTFDSGGYNIKTGPGCSIELMKFDMG 363
GVAAASANPP FIHL YKPP+G V VKLALVGKGLTFDSGGYNIKTGPGC IELMKFDMG
Sbjct: 938 GVAAASANPPHFIHLCYKPPTGPVNVKLALVGKGLTFDSGGYNIKTGPGCLIELMKFDMG 1117
Query: 364 GSAAVLGAAKALGQIKPLGVEVHFIVAACENMISGTGMRPGDVVTASNGKTIEVNNTDAE 423
GSAAV GAAKALGQIKPLGVEVHFIVAACENMISGTGMRPGD+VTASNGKTIEVNNTDAE
Sbjct: 1118GSAAVFGAAKALGQIKPLGVEVHFIVAACENMISGTGMRPGDIVTASNGKTIEVNNTDAE 1297
Query: 424 GRLTLADALVYACNQGVEKIIDLATLTGACVVALGPSIAGVFSPNDELVKEVLEASEVSG 483
GRLTLADALVYACNQG EKI+DLATLTGAC++ LG SIAGVF+P+D+L KEV +ASE SG
Sbjct: 1298GRLTLADALVYACNQGAEKIVDLATLTGACIIGLGSSIAGVFTPSDDLAKEVFDASEASG 1477
Query: 484 EKLWRLPIEESYWESMKSGVADMVNTGGRPGGSITAALFLKQFVDEKVQWLHIDMAGPVW 543
EKLWRLP+EESYWESMKSGVADMVNTGGRPG +ITAALFLKQFVDEKVQW+HIDMAGPVW
Sbjct: 1478EKLWRLPLEESYWESMKSGVADMVNTGGRPGSAITAALFLKQFVDEKVQWMHIDMAGPVW 1657
Query: 544 NDKKRSATGFGVSTLVEWVLKNSS 567
N+K+R ATGFGV+TLVEWVL+N+S
Sbjct: 1658NEKQRCATGFGVATLVEWVLRNAS 1729
>TC205472 similar to UP|Q43034 (Q43034) Leucine aminopeptidase (Cytosol
aminopeptidase) (Fragment) , partial (66%)
Length = 947
Score = 362 bits (929), Expect = e-100
Identities = 175/195 (89%), Positives = 188/195 (95%)
Frame = +2
Query: 373 KALGQIKPLGVEVHFIVAACENMISGTGMRPGDVVTASNGKTIEVNNTDAEGRLTLADAL 432
KALGQIK LGVEVHFIVAACENMISGT MRPGD+VT SNGKTIEVNNTDAEGRLTLADAL
Sbjct: 5 KALGQIKTLGVEVHFIVAACENMISGTSMRPGDIVTTSNGKTIEVNNTDAEGRLTLADAL 184
Query: 433 VYACNQGVEKIIDLATLTGACVVALGPSIAGVFSPNDELVKEVLEASEVSGEKLWRLPIE 492
VYACNQGVEKIIDLATLTGACVVALGPSIAGVF+P+D+L KEV EAS+VSGEKLWRLP+E
Sbjct: 185 VYACNQGVEKIIDLATLTGACVVALGPSIAGVFTPSDDLAKEVFEASDVSGEKLWRLPLE 364
Query: 493 ESYWESMKSGVADMVNTGGRPGGSITAALFLKQFVDEKVQWLHIDMAGPVWNDKKRSATG 552
ESYWE+MKSGVADM+NTGGR GG+I AALFLKQFVDEKVQW+HIDMAGPVWNDKKRSATG
Sbjct: 365 ESYWETMKSGVADMLNTGGRQGGAIVAALFLKQFVDEKVQWMHIDMAGPVWNDKKRSATG 544
Query: 553 FGVSTLVEWVLKNSS 567
FG++TLVEWVLKN+S
Sbjct: 545 FGIATLVEWVLKNAS 589
>TC207492 similar to UP|Q944P7 (Q944P7) AT4g30920/F6I18_170, partial (34%)
Length = 625
Score = 325 bits (832), Expect = 4e-89
Identities = 170/207 (82%), Positives = 186/207 (89%)
Frame = +3
Query: 132 VVRIKGLGSKRVGLIGLGQLPSTTALYKGLGEAVVAVAKSAQASNVAIVLASSEGLSSES 191
V+RI GLGSKRVGLIGLGQ PST + +KGLGEAVV AKS+QAS+VA+VLASSEGLS++S
Sbjct: 3 VLRITGLGSKRVGLIGLGQSPSTPSPFKGLGEAVVEAAKSSQASSVAVVLASSEGLSAQS 182
Query: 192 KLSTAYAIASGAVLGLFEDQRYKSESKKPAVRSIDIIGLGTGPDLEKKLKYAGDVSFGII 251
KLSTAYAIASGAVLGLFED RYKSE+KK +RSIDIIGLG GP+LEKKLKYAGDVS GII
Sbjct: 183 KLSTAYAIASGAVLGLFEDNRYKSEAKKSTLRSIDIIGLGMGPELEKKLKYAGDVSSGII 362
Query: 252 FGRELVNSPANVLTPGVLAEEASKVASTYSDVFTAKILDADQCKELKMGSYLGVAAASAN 311
FGRELVNSPANVLTPGVLAEEA+K+ASTYSDVFTAKIL+A+QC ELKMGSYLGVAAAS N
Sbjct: 363 FGRELVNSPANVLTPGVLAEEAAKIASTYSDVFTAKILNAEQCAELKMGSYLGVAAASEN 542
Query: 312 PPRFIHLTYKPPSGSVKVKLALVGKGL 338
PP FIHL YKP SG V VKLA +GL
Sbjct: 543 PPHFIHLCYKPLSGPVNVKLAFSWQGL 623
>BI970813 similar to PIR|T14912|T14 leucyl aminopeptidase (EC 3.4.11.1) -
parsley (fragment), partial (50%)
Length = 467
Score = 172 bits (437), Expect(2) = 1e-65
Identities = 85/100 (85%), Positives = 87/100 (87%)
Frame = -2
Query: 289 LDADQCKELKMGSYLGVAAASANPPRFIHLTYKPPSGSVKVKLALVGKGLTFDSGGYNIK 348
LDA+QCKELKMGSY+GVAAA ANP FIHL YKPP G V VKLAL GKGLTFDSGGY IK
Sbjct: 466 LDAEQCKELKMGSYVGVAAAXANPHHFIHLCYKPPPGPVNVKLALXGKGLTFDSGGYTIK 287
Query: 349 TGPGCSIELMKFDMGGSAAVLGAAKALGQIKPLGVEVHFI 388
TGPGC IELMKFDMGGSAAV GAAKALGQ PLGVEVHFI
Sbjct: 286 TGPGCLIELMKFDMGGSAAVFGAAKALGQXXPLGVEVHFI 167
Score = 96.3 bits (238), Expect(2) = 1e-65
Identities = 48/59 (81%), Positives = 52/59 (87%), Gaps = 2/59 (3%)
Frame = -3
Query: 383 VEVHFIV--AACENMISGTGMRPGDVVTASNGKTIEVNNTDAEGRLTLADALVYACNQG 439
+E FI+ ACENMISGTGMRPGD+VTASNGKTIEVN+TDAEGR TLADALVYAC QG
Sbjct: 189 LEWRFIL*PXACENMISGTGMRPGDIVTASNGKTIEVNSTDAEGRFTLADALVYACXQG 13
>TC207491 similar to UP|O65557 (O65557) Leucyl aminopeptidase-like protein,
partial (27%)
Length = 659
Score = 238 bits (607), Expect = 5e-63
Identities = 136/204 (66%), Positives = 152/204 (73%), Gaps = 2/204 (0%)
Frame = +3
Query: 22 LTSRIRFASFPKRSFHSTTKLMSQS-RYTTLGLTHPTNIEAPKISFSATDVDVTEWKGDI 80
LTSR+ FA+FP RS T KLMS + TLGLTHP+NIE PKISF A DVDVTEWKGDI
Sbjct: 54 LTSRLHFAAFPLRS--PTRKLMSHTLARATLGLTHPSNIETPKISFGAKDVDVTEWKGDI 227
Query: 81 LAVGVTEKDLTRDAKSRFENSILNKIDLKLDGLLSEASSEEDFSGKVGQSTVVRIKGLGS 140
LAVGVTEKDL RDA S+FEN IL+KID KL GLL+EASSEEDFSGKVGQSTV+RI GLGS
Sbjct: 228 LAVGVTEKDLARDANSKFENVILSKIDSKLGGLLAEASSEEDFSGKVGQSTVLRITGLGS 407
Query: 141 KRVGLIGLGQLPSTTALYKGLGEAVVAVAKSAQASNVAIVLASSEGLSSESKLSTAYAIA 200
KRVGLIGLGQ PST A +K LGEA+V AKS+QAS+VA+ LASSE L K
Sbjct: 408 KRVGLIGLGQSPSTPAPFKSLGEAIVEAAKSSQASSVAVALASSEXLFRTIKA*HCLCNC 587
Query: 201 S-GAVLGLFEDQRYKSESKKPAVR 223
G FED R KS +KK ++
Sbjct: 588 FWGCAWDYFEDNRSKSRTKKSTLK 659
>AW760437 similar to GP|27463709|gb| neutral leucine aminopeptidase
preprotein; preLAP-N; metallo-exopeptidase; leucyl
aminopeptidase;, partial (13%)
Length = 310
Score = 142 bits (358), Expect = 4e-34
Identities = 74/100 (74%), Positives = 81/100 (81%)
Frame = +2
Query: 368 VLGAAKALGQIKPLGVEVHFIVAACENMISGTGMRPGDVVTASNGKTIEVNNTDAEGRLT 427
VLGAAKALGQ+KPLG EVHFIVAA ENMISGTGMRPGD+ TASNGKTI+VNNTDAEGRLT
Sbjct: 11 VLGAAKALGQVKPLGAEVHFIVAA*ENMISGTGMRPGDIGTASNGKTIKVNNTDAEGRLT 190
Query: 428 LADALVYACNQGVEKIIDLATLTGACVVALGPSIAGVFSP 467
LA+ALVYA NQ EKII T T +V L PS + +P
Sbjct: 191 LANALVYASNQCCEKIIYSPTFTST*IVTLTPSTTNILTP 310
>TC213885 weakly similar to UP|Q9VUB7 (Q9VUB7) CG32133-PA, partial (4%)
Length = 540
Score = 29.6 bits (65), Expect = 3.8
Identities = 17/48 (35%), Positives = 29/48 (60%)
Frame = -2
Query: 164 AVVAVAKSAQASNVAIVLASSEGLSSESKLSTAYAIASGAVLGLFEDQ 211
AV AVA +A+A+ VA+V+ E ++ + A A A+GAV + ++
Sbjct: 302 AVAAVAATAEAAAVAVVVGVVEVEAAAAVAGVAEAAAAGAVAAVVAEE 159
>TC219543 weakly similar to UP|Q94CH8 (Q94CH8) Family II lipase EXL1, partial
(49%)
Length = 1085
Score = 29.6 bits (65), Expect = 3.8
Identities = 34/142 (23%), Positives = 60/142 (41%), Gaps = 8/142 (5%)
Frame = +2
Query: 75 EWKGDILAVGVTEKDLTRDAKSRF-----ENSILNKIDLKLDGLLSEASSEEDFSGKVGQ 129
E+K I+ + + T +KS + N I N ++ G + + D
Sbjct: 452 EYKNKIMEIVGENRTATIISKSIYILCTGSNDITNTYFVR--GGEYDIQAYTDLMASQAT 625
Query: 130 STVVRIKGLGSKRVGLIG---LGQLPSTTALYKGLGEAVVAVAKSAQASNVAIVLASSEG 186
+ + + GLG++R+G++G LG +PS L+ G+ A + N A VL +S+
Sbjct: 626 NFLQELYGLGARRIGVVGLPVLGCVPSQRTLHGGIFRA------CSDFENEAAVLFNSKL 787
Query: 187 LSSESKLSTAYAIASGAVLGLF 208
S L + A L L+
Sbjct: 788 SSQMDALKKQFQEARFVYLDLY 853
>TC217055 similar to UP|O64418 (O64418) Aldehyde oxidase , partial (14%)
Length = 847
Score = 29.3 bits (64), Expect = 4.9
Identities = 14/44 (31%), Positives = 23/44 (51%)
Frame = +3
Query: 415 IEVNNTDAEGRLTLADALVYACNQGVEKIIDLATLTGACVVALG 458
+E++ + E R D ++Y C Q + +DL + GA V LG
Sbjct: 105 VEIDLLNGETRFLQTD-IIYDCGQSLNPAVDLGQIEGAFVQGLG 233
>TC227318 similar to UP|Q75H88 (Q75H88) Expressed protein, 3'-partial
(Fragment), partial (11%)
Length = 790
Score = 28.9 bits (63), Expect = 6.4
Identities = 13/33 (39%), Positives = 19/33 (57%)
Frame = +3
Query: 12 FSKSSSSSLFLTSRIRFASFPKRSFHSTTKLMS 44
++K SSS F +S F +F + FH T +MS
Sbjct: 198 YNKKPSSSSFFSSPRLFTNFTPKGFHETETMMS 296
>TC234722 similar to UP|Q6WAY9 (Q6WAY9) Pol (Fragment), partial (32%)
Length = 482
Score = 28.5 bits (62), Expect = 8.4
Identities = 12/38 (31%), Positives = 21/38 (54%)
Frame = -3
Query: 459 PSIAGVFSPNDELVKEVLEASEVSGEKLWRLPIEESYW 496
P G +++ +K VLE VS K W L +++++W
Sbjct: 228 PQTNGQAEVSNKEIKRVLENIVVSSRKDWALKLDDAFW 115
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.132 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,071,908
Number of Sequences: 63676
Number of extensions: 239295
Number of successful extensions: 923
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 914
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 921
length of query: 567
length of database: 12,639,632
effective HSP length: 102
effective length of query: 465
effective length of database: 6,144,680
effective search space: 2857276200
effective search space used: 2857276200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC136953.9


