

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136953.3 + phase: 0 /pseudo
(630 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
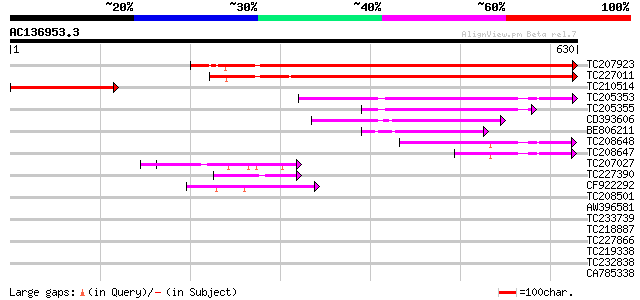
Score E
Sequences producing significant alignments: (bits) Value
TC207923 homologue to UP|Q9M0A0 (Q9M0A0) Signal recognition part... 703 0.0
TC227011 homologue to UP|Q9M0A0 (Q9M0A0) Signal recognition part... 693 0.0
TC210514 similar to UP|Q9M0A0 (Q9M0A0) Signal recognition partic... 221 6e-58
TC205353 homologue to UP|SR52_LYCES (P49972) Signal recognition ... 132 4e-31
TC205355 homologue to UP|SR52_LYCES (P49972) Signal recognition ... 103 2e-22
CD393606 similar to GP|3746964|gb| signal recognition particle 5... 99 6e-21
BE806211 similar to GP|29897491|gb Cell division protein ftsY {B... 94 2e-19
TC208648 similar to PIR|T52612|T52612 SRP receptor homolog FtsY ... 94 2e-19
TC208647 similar to PIR|T52612|T52612 SRP receptor homolog FtsY ... 56 4e-08
TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%) 48 2e-05
TC227390 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacety... 42 6e-04
CF922292 42 8e-04
TC208501 similar to GB|AAN31116.1|23506209|AY149962 At3g06130/F2... 40 0.002
AW396581 39 0.005
TC233739 37 0.035
TC218887 similar to UP|Q41042 (Q41042) Pisum sativum L. (clone n... 36 0.045
TC227866 similar to UP|P2B1_DROME (P48456) Serine/threonine prot... 36 0.059
TC219338 similar to UP|O64403 (O64403) Pasticcino 1-D, partial (... 35 0.077
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 35 0.13
CA785338 similar to GP|3850569|gb|A ESTs gb|T21276 gb|T45403 a... 34 0.17
>TC207923 homologue to UP|Q9M0A0 (Q9M0A0) Signal recognition particle
receptor-like protein (AT4g30600/F17I23_60), partial
(60%)
Length = 1569
Score = 703 bits (1815), Expect = 0.0
Identities = 378/436 (86%), Positives = 399/436 (90%), Gaps = 7/436 (1%)
Frame = +2
Query: 202 VNGKENDSVNNGAFDVNRLQKKVRNKGGNGKKTDAVV---TKAEP-KKVVKKNRVWDEKP 257
VNG+ N S N GAFDV++LQK +R KGG KKTD VV +KAEP KKV KKNRVWDE
Sbjct: 68 VNGRHNGSPNVGAFDVHKLQK-LRTKGG--KKTDTVVAKASKAEPNKKVTKKNRVWDEAA 238
Query: 258 V-ETKLDFTDHVDIDGDADKDRKVDYL-AKEQGESMMDKDEIFSSDSE-DEEDDDDDNAG 314
ETKLDFTDH DG+ R +D++ A +QGESMMDK+EI SS+SE +EED+D+++AG
Sbjct: 239 TTETKLDFTDHSGEDGE----RNIDFVVAADQGESMMDKEEIVSSESEQEEEDEDEEDAG 406
Query: 315 KKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVA 374
K KPDAK KGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVA
Sbjct: 407 KNRKPDAKSKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVA 586
Query: 375 ASLEGKKLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVN 434
ASLEGKKLASFTRISSTV AAME+AL+RILTPRRSIDILRDVHAAKEQRKPYVVVFVGVN
Sbjct: 587 ASLEGKKLASFTRISSTVHAAMEEALIRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVN 766
Query: 435 GVGKSTNLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAV 494
GVGKSTNLAKVAYWL QH V+VMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAV
Sbjct: 767 GVGKSTNLAKVAYWLLQHKVSVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAV 946
Query: 495 VAKEAIQEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAV 554
VAKEAIQEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAV
Sbjct: 947 VAKEAIQEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAV 1126
Query: 555 DQLSKFNQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSY 614
DQLSKFNQKLADL+TSP PRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSY
Sbjct: 1127DQLSKFNQKLADLATSPNPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSY 1306
Query: 615 TDLKKLNVKSIVKTLL 630
TDLKKLNVKSIVKTLL
Sbjct: 1307TDLKKLNVKSIVKTLL 1354
>TC227011 homologue to UP|Q9M0A0 (Q9M0A0) Signal recognition particle
receptor-like protein (AT4g30600/F17I23_60), partial
(55%)
Length = 1496
Score = 693 bits (1788), Expect = 0.0
Identities = 362/411 (88%), Positives = 377/411 (91%), Gaps = 3/411 (0%)
Frame = +3
Query: 223 KVRNKGGNGKKTDAVVT---KAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRK 279
K + GNGKK DA+V K EPKKV K NRVWD+ +TKLDFTDHVD DGD R
Sbjct: 3 KGKKGNGNGKKKDALVVAAAKGEPKKVDKPNRVWDQPAPQTKLDFTDHVDGDGD----RS 170
Query: 280 VDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAKKKGWFSSMFQSIAGKAN 339
D+LAKEQGESMMDK+EI SSDSE E+DDDD GK + P AKKKGWFSSMFQSIAGKAN
Sbjct: 171 ADFLAKEQGESMMDKEEILSSDSEVEDDDDD--TGKDNMPVAKKKGWFSSMFQSIAGKAN 344
Query: 340 LEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISSTVQAAMEDA 399
LEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISSTV AME+A
Sbjct: 345 LEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISSTVHTAMEEA 524
Query: 400 LVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQHNVNVMMA 459
LVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWL QHNV+VMMA
Sbjct: 525 LVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLLQHNVSVMMA 704
Query: 460 ACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEASRNGSDVVLVDTAGRM 519
ACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPA+VAKEAIQEA+RNGSDVVLVDTAGRM
Sbjct: 705 ACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAIVAKEAIQEAARNGSDVVLVDTAGRM 884
Query: 520 QDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKLADLSTSPTPRLIDGI 579
QDNEPLMRALSKL+YLNNPDL+LFVGEALVGNDAVDQLSKFNQKLADLSTSPTPRLIDGI
Sbjct: 885 QDNEPLMRALSKLVYLNNPDLILFVGEALVGNDAVDQLSKFNQKLADLSTSPTPRLIDGI 1064
Query: 580 LLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKLNVKSIVKTLL 630
LLTKFDTIDDKVGAALSMVY+SGAPVMFVGCGQSYTDLKKLNVKSI KTLL
Sbjct: 1065LLTKFDTIDDKVGAALSMVYVSGAPVMFVGCGQSYTDLKKLNVKSIAKTLL 1217
>TC210514 similar to UP|Q9M0A0 (Q9M0A0) Signal recognition particle
receptor-like protein (AT4g30600/F17I23_60), partial
(19%)
Length = 436
Score = 221 bits (564), Expect = 6e-58
Identities = 105/121 (86%), Positives = 116/121 (95%)
Frame = +1
Query: 1 MLEQLLIFTRGGLILWSCNEIGNALKGSPIDTLIRSCLLEERSGASSYNYDAPGAAYSLK 60
MLEQLLIFTRGGLILW+CN + NAL+GSPIDTLIRSCLLEERSGA+S+NYDAPGAAYSLK
Sbjct: 73 MLEQLLIFTRGGLILWTCNHLSNALRGSPIDTLIRSCLLEERSGAASFNYDAPGAAYSLK 252
Query: 61 WTFHNDLGLVFVAVYQRILHLLYVDDLLAAVKREFSQVYDPTRTVYRDFDEIFKQLKIEA 120
WTFHNDLGLVFVAVYQR+LHLLYVDDLLAAVKREFS++Y P +T YRDFDEIF+QL+IEA
Sbjct: 253 WTFHNDLGLVFVAVYQRVLHLLYVDDLLAAVKREFSRLYHPQKTAYRDFDEIFQQLQIEA 432
Query: 121 E 121
E
Sbjct: 433 E 435
>TC205353 homologue to UP|SR52_LYCES (P49972) Signal recognition particle 54
kDa protein 2 (SRP54), complete
Length = 1889
Score = 132 bits (333), Expect = 4e-31
Identities = 86/310 (27%), Positives = 150/310 (47%)
Frame = +3
Query: 321 AKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGK 380
A+ G S Q ++ +++ L L + L+ +V ++ L ++ +
Sbjct: 75 AELGGSISRALQQMSNATVIDEKVLNDCLNVITRALLQSDVQFKLVRDLQTNIKNIVNLD 254
Query: 381 KLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKST 440
LA+ +Q A+ + L +IL P + ++ K VV+FVG+ G GK+T
Sbjct: 255 DLAAGHNKRRIIQQAVFNELCKILDPGKP-------SFTPKKGKTSVVMFVGLQGSGKTT 413
Query: 441 NLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAI 500
K AY+ Q+ + DTFR+GA +QL+ +A + +IP + E DP +A E +
Sbjct: 414 TCTKYAYYHQKKGWKPALVCADTFRAGAFDQLKQNATKAKIPFYGSYMESDPVKIAVEGV 593
Query: 501 QEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKF 560
+ + D+++VDT+GR + L + ++ PDLV+FV ++ +G A DQ F
Sbjct: 594 ERFKKENCDLIIVDTSGRHKQEAALFEEMRQVSEATKPDLVIFVMDSSIGQAAFDQAQAF 773
Query: 561 NQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKL 620
Q +A + +++TK D K G ALS V + +PV+F+G G+ + +
Sbjct: 774 KQSVA----------VGAVIVTKMDG-HAKGGGALSAVAATKSPVIFIGTGEHMDEFEVF 920
Query: 621 NVKSIVKTLL 630
+VK V LL
Sbjct: 921 DVKPFVSRLL 950
>TC205355 homologue to UP|SR52_LYCES (P49972) Signal recognition particle 54
kDa protein 2 (SRP54), partial (37%)
Length = 554
Score = 103 bits (257), Expect = 2e-22
Identities = 58/194 (29%), Positives = 100/194 (50%)
Frame = +3
Query: 392 VQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQ 451
+Q A+ + L +IL P + ++ K VV+FVG+ G GK+T K A++ Q+
Sbjct: 21 IQQAVFNELCKILDPGKP-------SFTPKKGKTSVVMFVGLQGSGKTTTCTKYAFYHQK 179
Query: 452 HNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEASRNGSDVV 511
+ DTFR+GA +QL+ +A + +IP + E DP +A E ++ + D++
Sbjct: 180 KGWKPALVCADTFRAGAFDQLKQNATKAKIPFYGSYMESDPVKIAVEGVERFKQENCDLI 359
Query: 512 LVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKLADLSTSP 571
+VDT+GR + L + ++ PDLV+FV ++ +G A DQ F Q +A
Sbjct: 360 IVDTSGRHKQEAALFEEMRQVSEATKPDLVIFVMDSSIGQAAFDQAQAFKQSVA------ 521
Query: 572 TPRLIDGILLTKFD 585
+ +++TK D
Sbjct: 522 ----VGAVIVTKMD 551
>CD393606 similar to GP|3746964|gb| signal recognition particle 54 kDa
subunit precursor {Arabidopsis thaliana}, partial (37%)
Length = 630
Score = 99.0 bits (245), Expect = 6e-21
Identities = 59/215 (27%), Positives = 117/215 (53%)
Frame = +3
Query: 336 GKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISSTVQAA 395
G+ L K ++ ++ ++ L+ +V+ + + +SV+ G + R +
Sbjct: 3 GEEVLSKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVGVGVIRGVRPDQQLVKI 182
Query: 396 MEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQHNVN 455
+ + LV+++ S ++ AK P V++ G+ GVGK+T AK+A +L++ +
Sbjct: 183 VHEELVQLMGGEVS-----ELVFAKSG--PTVILLAGLQGVGKTTVCAKLANYLKKQGKS 341
Query: 456 VMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEASRNGSDVVLVDT 515
M+ A D +R A++QL +++ +P++ G + P+ +AK+ ++EA + DVV+VDT
Sbjct: 342 CMLVAGDVYRPAAIDQLAILGKQVDVPVYTAGTDVKPSEIAKQGLEEAKKKKIDVVIVDT 521
Query: 516 AGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVG 550
AGR+Q ++ +M L ++ NP VL V +A+ G
Sbjct: 522 AGRLQIDKTMMDELKEVKKALNPTEVLLVVDAMTG 626
>BE806211 similar to GP|29897491|gb Cell division protein ftsY {Bacillus
cereus ATCC 14579}, partial (43%)
Length = 436
Score = 93.6 bits (231), Expect = 2e-19
Identities = 54/141 (38%), Positives = 83/141 (58%)
Frame = -1
Query: 392 VQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQ 451
V A + L++I T + D +++ K+ V+FVGVNGVGK+T + K+A+
Sbjct: 421 VPAVISKKLIKIYTG--TSDFTNEINEPKDGSTD--VLFVGVNGVGKTTTIGKMAHKF*S 254
Query: 452 HNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEASRNGSDVV 511
+V++AA DTFR GA+EQL R+ + + ++G DPA V +A+Q A DV+
Sbjct: 253 EGKSVLLAAGDTFRDGAIEQL*VSGDRVGVEVIKQGSGSDPAAVMYDAVQAAKARKVDVL 74
Query: 512 LVDTAGRMQDNEPLMRALSKL 532
L DTAGR+Q+ LM+ L K+
Sbjct: 73 LCDTAGRLQNKVNLMKELDKV 11
>TC208648 similar to PIR|T52612|T52612 SRP receptor homolog FtsY precursor,
chloroplast [validated] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (53%)
Length = 845
Score = 93.6 bits (231), Expect = 2e-19
Identities = 66/203 (32%), Positives = 100/203 (48%), Gaps = 7/203 (3%)
Frame = +1
Query: 434 NGVGKSTNLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPA 493
NG GK+T+L K+AY L+ ++MAA DTFR+ A +QL A R I EK A
Sbjct: 4 NGGGKTTSLGKLAYRLKNEGAKILMAAGDTFRAAASDQLEIWAGRTGCEIVVAESEKAKA 183
Query: 494 -VVAKEAIQEASRNGSDVVLVDTAGRMQDNEPLMRALSKL------IYLNNPDLVLFVGE 546
V +A+++ G D+VL DT+GR+ N LM L + P+ +L V +
Sbjct: 184 SSVLSQAVKKGKELGFDIVLCDTSGRLHTNYSLMEELISCKKSVAKVVPGAPNEILLVLD 363
Query: 547 ALVGNDAVDQLSKFNQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVM 606
G + + Q +FN + + G++LTK D + G +S+V G PV
Sbjct: 364 GTTGLNMLPQAREFNDVVG----------VTGLILTKLDG-SARGGCVVSVVDELGIPVK 510
Query: 607 FVGCGQSYTDLKKLNVKSIVKTL 629
FVG G+ DL+ + +S V +
Sbjct: 511 FVGVGEGVEDLQPFDAESFVNAI 579
>TC208647 similar to PIR|T52612|T52612 SRP receptor homolog FtsY precursor,
chloroplast [validated] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (37%)
Length = 817
Score = 56.2 bits (134), Expect = 4e-08
Identities = 39/141 (27%), Positives = 66/141 (46%), Gaps = 6/141 (4%)
Frame = +1
Query: 495 VAKEAIQEASRNGSDVVLVDTAGRMQDNEPLMRALSKL------IYLNNPDLVLFVGEAL 548
V +A+++ G D+VL DT+GR+ N LM L + P+ +L V +
Sbjct: 13 VLSQAVKKGKELGFDIVLCDTSGRLHTNYSLMEELISCKKSVAKVVPGAPNEILLVLDGT 192
Query: 549 VGNDAVDQLSKFNQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFV 608
G + + Q +FN + + G++LTK D + G +S+V G PV FV
Sbjct: 193 TGLNMLPQAREFNDVVG----------VTGLVLTKLDG-SARGGCVVSVVDELGIPVKFV 339
Query: 609 GCGQSYTDLKKLNVKSIVKTL 629
G G+ DL+ + ++ V +
Sbjct: 340 GVGEGVEDLQPFDAEAFVNAI 402
>TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%)
Length = 1005
Score = 47.8 bits (112), Expect = 2e-05
Identities = 50/181 (27%), Positives = 73/181 (39%), Gaps = 20/181 (11%)
Frame = +3
Query: 164 NDGDGKNGKKNSENDRSAIVNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFDVNRLQKK 223
+DG+ K K E + + G+ ++ V GKEND G D + +KK
Sbjct: 6 DDGEEKKEKDKKEKKKKENKDKGDKTDVEK--------VKGKEND----GEDDEEKKEKK 149
Query: 224 VRNKGGNGKKTDAVVTKA---------EPKKVVKKNRVWDEKPVETKLDF---------T 265
+ K KTDA K E K+ KK D V+ K D
Sbjct: 150 KKEKKDKDGKTDAEKIKKKEDDGKDDEEKKEKKKKYDKIDAXKVKGKEDDGKDEGNKEKK 329
Query: 266 DHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDD--DDNAGKKSKPDAKK 323
D DGD ++ ++ KE+ + DKDE + E ++D+ DD KK K D K+
Sbjct: 330 DKEKGDGDGEEKKEKKDKEKEKKKEKKDKDEETDTLKEKGKNDEGEDDEGNKKKKKDKKE 509
Query: 324 K 324
K
Sbjct: 510 K 512
Score = 45.4 bits (106), Expect = 7e-05
Identities = 47/197 (23%), Positives = 82/197 (40%), Gaps = 18/197 (9%)
Frame = +3
Query: 146 KGDGSDGKKNGSAGGGLKNDGDGK-NGKKNSENDRSAIVNNGNGYNLRSNGVVGNVSVNG 204
+ DG D ++ K D DGK + +K + + + + + V G
Sbjct: 108 ENDGEDDEEKKEKKKKEKKDKDGKTDAEKIKKKEDDGKDDEEKKEKKKKYDKIDAXKVKG 287
Query: 205 KENDSVNNGAFDVNRLQKKVRNKG-GNGKKTDAVVTKAEPKKVVKKNRVWDEKPVETKLD 263
KE+D + G +KK + KG G+G++ K + KK KK++ + ++ K
Sbjct: 288 KEDDGKDEG-----NKEKKDKEKGDGDGEEKKEKKDKEKEKKKEKKDKDEETDTLKEKGK 452
Query: 264 FTDHVDIDG---------DADKDRKVDYLAKEQGESMMDKDEIFSSD-------SEDEED 307
+ D +G + +KD K + KE+GE K E+ D E E++
Sbjct: 453 NDEGEDDEGNKKKKKDKKEKEKDHKKEKKDKEEGEKEDSKVEVSVRDIDIEEIKKEGEKE 632
Query: 308 DDDDNAGKKSKPDAKKK 324
D + GK+ K KK+
Sbjct: 633 DKGKDGGKEVKEKKKKE 683
>TC227390 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (24%)
Length = 808
Score = 42.4 bits (98), Expect = 6e-04
Identities = 37/102 (36%), Positives = 45/102 (43%), Gaps = 4/102 (3%)
Frame = +1
Query: 227 KGGNGK-KTDAVVTKA-EPKKVVKKNRVWDE--KPVETKLDFTDHVDIDGDADKDRKVDY 282
K NGK +T A K E KK V K + K VE K D + D D D D D
Sbjct: 49 KAENGKPETKAEELKVPESKKAVAKASASAKQVKIVEPKKDNEEDSDDDSDEDDD----- 213
Query: 283 LAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAKKK 324
E M D D +S+D+EDDD++ K K D KK
Sbjct: 214 -FGSSDEEMEDADSDSDDESDDKEDDDEETHPKNKKADLGKK 336
>CF922292
Length = 722
Score = 42.0 bits (97), Expect = 8e-04
Identities = 43/161 (26%), Positives = 67/161 (40%), Gaps = 13/161 (8%)
Frame = -2
Query: 197 VGNVSVNGKENDSVNNGAFDV--NRLQKKVRNKG--------GNGKKTDAVVTKAEPKKV 246
V NV V+ + N V A D N Q+ N+G G+G T + + K+
Sbjct: 490 VQNVDVSQENNSEVKEQATDPSNNNSQQFEDNRGDLSEDATKGDGSVTPDKNSDVKEKQE 311
Query: 247 VKKNRVWDEKPVE---TKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSE 303
K + EKP E T+ T + D+D+ ++ + Q +S D+ E S +E
Sbjct: 310 EKSDEKSQEKPSEDTKTENQDTSVSEKRSDSDESQQKSDSDESQQKSDSDESEKKSDSAE 131
Query: 304 DEEDDDDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEKSD 344
E+ D D + KKS D +K S+ + KSD
Sbjct: 130 SEKKSDSDESEKKSDSDETEKSSESNDNKQFDSDERENKSD 8
>TC208501 similar to GB|AAN31116.1|23506209|AY149962 At3g06130/F28L1_7
{Arabidopsis thaliana;} , partial (26%)
Length = 1081
Score = 40.4 bits (93), Expect = 0.002
Identities = 32/96 (33%), Positives = 40/96 (41%), Gaps = 1/96 (1%)
Frame = +3
Query: 136 GGNR-KQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKKNSENDRSAIVNNGNGYNLRSN 194
GGN K K +G G N + GGG KN+G GKNG E NNG G + +N
Sbjct: 411 GGNEGKNGNGGKKEGGGGGNNQTQGGGNKNNG-GKNGGGVPEGKNGNNKNNGGGGGIPNN 587
Query: 195 GVVGNVSVNGKENDSVNNGAFDVNRLQKKVRNKGGN 230
NG NNG + + N GG+
Sbjct: 588 --------NGNGGKKGNNGMAEGVQAMNMFPNMGGH 671
Score = 28.9 bits (63), Expect = 7.2
Identities = 24/99 (24%), Positives = 30/99 (30%), Gaps = 14/99 (14%)
Frame = +3
Query: 147 GDGSDGKKNGSAGGGLKNDGDGKNGKKNSENDRSAIV------------NNGNGYNLRSN 194
G+G+ N +G G NGKK + NGNG
Sbjct: 279 GNGAQMMSNSMMNAQKGGNGGGNNGKKGGGGSGGPVPVQVHNMGGGNEGKNGNGGKKEGG 458
Query: 195 GVVGNVSVNG--KENDSVNNGAFDVNRLQKKVRNKGGNG 231
G N + G K N N G + N GG G
Sbjct: 459 GGGNNQTQGGGNKNNGGKNGGGVPEGKNGNNKNNGGGGG 575
>AW396581
Length = 401
Score = 39.3 bits (90), Expect = 0.005
Identities = 27/104 (25%), Positives = 45/104 (42%)
Frame = -1
Query: 221 QKKVRNKGGNGKKTDAVVTKAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKV 280
+KK + K KTD K + K KK K + K D TD + G D
Sbjct: 329 EKKKKEKKDKDDKTDVEKVKGKEDKEKKK------KEKKVKDDKTDAEKVKGKEDDGEDN 168
Query: 281 DYLAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAKKK 324
+ +++ + +KD+ + ++DD ++ GKK K +KK
Sbjct: 167 EEKEEKKKKKKKEKDDKIDVEKVKGKEDDGEDEGKKEKKKKEKK 36
Score = 35.4 bits (80), Expect = 0.077
Identities = 27/84 (32%), Positives = 39/84 (46%)
Frame = -1
Query: 241 AEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSS 300
AE KK KK K + K D TD + G DK++K + E + D+ +
Sbjct: 344 AEEKKEKKK------KEKKDKDDKTDVEKVKGKEDKEKK-------KKEKKVKDDKTDAE 204
Query: 301 DSEDEEDDDDDNAGKKSKPDAKKK 324
+ +EDD +DN K+ K KKK
Sbjct: 203 KVKGKEDDGEDNEEKEEKKKKKKK 132
>TC233739
Length = 562
Score = 36.6 bits (83), Expect = 0.035
Identities = 16/26 (61%), Positives = 19/26 (72%)
Frame = +3
Query: 300 SDSEDEEDDDDDNAGKKSKPDAKKKG 325
SD+EDEE++DD N KK D KKKG
Sbjct: 45 SDNEDEEEEDDKNKEKKDGEDEKKKG 122
>TC218887 similar to UP|Q41042 (Q41042) Pisum sativum L. (clone na-481-5),
partial (13%)
Length = 559
Score = 36.2 bits (82), Expect = 0.045
Identities = 31/112 (27%), Positives = 52/112 (45%), Gaps = 10/112 (8%)
Frame = +1
Query: 223 KVRNKGGNGKKTDAVVTKAEPKK-----VVKKNRVWDEKPVETKLDFTDHVDIDGDADKD 277
K KGG + D + + KK V +K + E V+ K + + + + +K
Sbjct: 211 KSAKKGGKRQAQDEIEKQVSAKKQKIEEVAQKQK--KEAKVQKKKESSSDDSSESEEEKP 384
Query: 278 RKVDYLAKE----QGESMMDKDEIFSSDSEDEEDDDDD-NAGKKSKPDAKKK 324
L+K+ G ++ KD+ SS SED+ D+D+ NA K K A+K+
Sbjct: 385 APKPVLSKKVPAKNGAALPKKDKPVSSSSEDDSSDEDEVNAKKPHKVVAEKQ 540
>TC227866 similar to UP|P2B1_DROME (P48456) Serine/threonine protein
phosphatase 2B catalytic subunit 1
(Calmodulin-dependent calcineurin A1 subunit) , partial
(3%)
Length = 1013
Score = 35.8 bits (81), Expect = 0.059
Identities = 29/94 (30%), Positives = 37/94 (38%), Gaps = 22/94 (23%)
Frame = +3
Query: 136 GGNRKQQVTWKGDGSDGKKNGSAGGG---------------------LKNDGDGKNGKKN 174
GG T K + GKK+G GGG KN G NGK N
Sbjct: 114 GGGSADTATNKNKNNKGKKDGGGGGGGGGKFDKIDFDFQDFDIPPTKGKNGKGGNNGKNN 293
Query: 175 SENDRSAIVNNGN-GYNLRSNGVVGNVSVNGKEN 207
ND+ NNGN G+++ G +G + G N
Sbjct: 294 --NDKGHGSNNGNVGHHMGPMGQMGPMVQRGPMN 389
>TC219338 similar to UP|O64403 (O64403) Pasticcino 1-D, partial (17%)
Length = 758
Score = 35.4 bits (80), Expect = 0.077
Identities = 36/130 (27%), Positives = 52/130 (39%), Gaps = 9/130 (6%)
Frame = +1
Query: 213 GAFDVNRLQKKVRNKGGNGKKTDAVVT----KAEPKKVVKKNR-----VWDEKPVETKLD 263
G F+ R KV K ++DA K + + V KK R ++D+KP E
Sbjct: 139 GDFEEARADFKVMMKVDKSTESDATAALQKLKQKEQDVEKKARKQFKGLFDKKPGEIS-- 312
Query: 264 FTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAKK 323
D+DGD Q S KD+ DS+ +D A P+A++
Sbjct: 313 -EAKADVDGD-------------QITSESQKDDEVHGDSDGTNSEDSHEA----PPEAQR 438
Query: 324 KGWFSSMFQS 333
GWFS + S
Sbjct: 439 TGWFSLFWPS 468
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 34.7 bits (78), Expect = 0.13
Identities = 27/94 (28%), Positives = 45/94 (47%), Gaps = 4/94 (4%)
Frame = +2
Query: 225 RNKGGNGKKTDAVVTKAEPKKVVKKNRVW--DEKPVETKLDFTDHVDIDGDADKDRKVDY 282
R + GK+ + + K ++N +W D K TK + ++ V+ + +V
Sbjct: 122 RCRATGGKRREEAAQRIS-KSRGERNVLWFLDSKMKATK-ELSEQVE-------ENEVKV 274
Query: 283 LAKEQGESMMDKDEIF--SSDSEDEEDDDDDNAG 314
L +E E DE+ + D EDEE+DDDD+ G
Sbjct: 275 LVEEDDEEYESVDEVVDDAEDDEDEEEDDDDDEG 376
>CA785338 similar to GP|3850569|gb|A ESTs gb|T21276 gb|T45403 and
gb|AA586113 come from this gene. {Arabidopsis thaliana},
partial (10%)
Length = 435
Score = 34.3 bits (77), Expect = 0.17
Identities = 30/115 (26%), Positives = 52/115 (45%), Gaps = 6/115 (5%)
Frame = +2
Query: 199 NVSVNGKENDSVNNGAFDVNRLQKKV------RNKGGNGKKTDAVVTKAEPKKVVKKNRV 252
NV+ N EN S++ D +K+ + G+ K +D V ++P +++
Sbjct: 62 NVASNVPENVSISGDKLDAKYEEKEASEGKIDNDNNGSCKDSDISVVSSQPSMPGEEDIG 241
Query: 253 WDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEED 307
WDE +E ++ D DG R + + S+ D+DE S D ED++D
Sbjct: 242 WDE--IED-IESNDESKGDGVGSASR----IDLRKRLSVADQDEDLSWDIEDDDD 385
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.133 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,338,669
Number of Sequences: 63676
Number of extensions: 246668
Number of successful extensions: 2284
Number of sequences better than 10.0: 136
Number of HSP's better than 10.0 without gapping: 1649
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1932
length of query: 630
length of database: 12,639,632
effective HSP length: 103
effective length of query: 527
effective length of database: 6,081,004
effective search space: 3204689108
effective search space used: 3204689108
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC136953.3


