

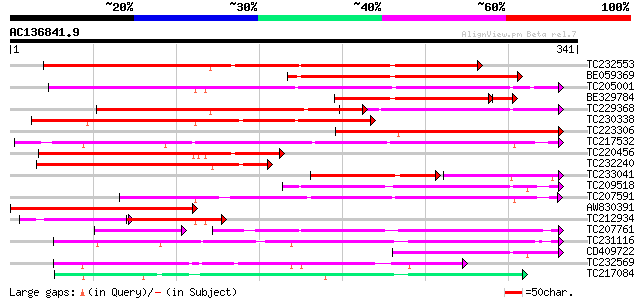
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.9 + phase: 0
(341 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC232553 similar to UP|Q9FXE5 (Q9FXE5) F12A21.4, partial (59%) 236 8e-63
BE059369 weakly similar to PIR|T09416|T094 coil protein PO22 mi... 213 8e-56
TC205001 similar to UP|O82681 (O82681) Lanatoside 15'-O-acetyles... 184 7e-47
BE329784 157 7e-43
TC229368 similar to PIR|T48618|T48618 early nodule-specific prot... 164 5e-41
TC230338 similar to UP|Q8W0Y6 (Q8W0Y6) Enod8.2, partial (49%) 163 1e-40
TC223306 weakly similar to PIR|T48618|T48618 early nodule-specif... 129 1e-30
TC217532 weakly similar to UP|Q9LHW8 (Q9LHW8) ESTs AU075812(E608... 125 4e-29
TC220456 weakly similar to UP|O81262 (O81262) Early nodule-speci... 125 4e-29
TC232240 weakly similar to UP|Q9FXE5 (Q9FXE5) F12A21.4, partial ... 113 1e-25
TC233041 weakly similar to UP|Q8W0Y7 (Q8W0Y7) Enod8.3 (Fragment)... 75 1e-25
TC209518 weakly similar to UP|O82076 (O82076) Lipase homolog (Fr... 112 2e-25
TC207591 weakly similar to UP|Q9LHW8 (Q9LHW8) ESTs AU075812(E608... 97 8e-21
AW830391 96 3e-20
TC212934 similar to UP|O82681 (O82681) Lanatoside 15'-O-acetyles... 62 1e-19
TC207761 weakly similar to UP|Q8L8G1 (Q8L8G1) Lipase SIL1, parti... 68 2e-18
TC231116 similar to UP|Q9FFC6 (Q9FFC6) GDSL-motif lipase/hydrola... 87 1e-17
CD409722 81 6e-16
TC232569 77 9e-15
TC217084 76 2e-14
>TC232553 similar to UP|Q9FXE5 (Q9FXE5) F12A21.4, partial (59%)
Length = 832
Score = 236 bits (603), Expect = 8e-63
Identities = 125/272 (45%), Positives = 174/272 (63%), Gaps = 8/272 (2%)
Frame = +2
Query: 21 SKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFISEELE 80
SK+C +PAI+NFGDSNSDTG A P+G S+F +GR CDGRLI+DF++++L
Sbjct: 20 SKQCHFPAIFNFGDSNSDTGGLSAAFGQAGPPHGESYFHHPAGRYCDGRLIVDFLAKKLG 199
Query: 81 LPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLG--------FQVSQFILFKSHT 132
LPYLS++L+SVGSNY HGANFA A + IRP L G Q +QF F+ T
Sbjct: 200 LPYLSAFLDSVGSNYSHGANFATAGSTIRPQNTTLHQTGGFSPFSLDVQFNQFSDFQRRT 379
Query: 133 KILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQ 192
+ F ++ +P+ EDFS+A+YT DIGQND+ G N S ++V+ +PD+L+Q
Sbjct: 380 Q--FFHNKGGVYKTLLPKAEDFSQALYTFDIGQNDLASGYFH-NMSTDQVKAYVPDVLAQ 550
Query: 193 FTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYN 252
F ++ +YN R FW+HNTGP+ C+PY +P K +D C P+NE+A+ +N
Sbjct: 551 FKNVIKYVYNHGGRSFWVHNTGPVGCLPYIMDLHPVK--PSLVDKAXCATPYNEVAKFFN 724
Query: 253 RQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLI 284
+LK+ V QL + PLA TYVDVY+VK++LI
Sbjct: 725 SKLKEVVVQLTKELPLAAITYVDVYSVKHSLI 820
>BE059369 weakly similar to PIR|T09416|T094 coil protein PO22
microspore/pollen-specific - alfalfa, partial (15%)
Length = 411
Score = 213 bits (543), Expect = 8e-56
Identities = 102/141 (72%), Positives = 119/141 (84%)
Frame = +1
Query: 168 IGYGLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYP 227
+ +GLQ ++S+E+V +SIP+IL+QF QAVQ+LYN ARVFWIHNTGPI C+PY Y +Y
Sbjct: 1 LAFGLQ--HTSQEQVIKSIPEILNQFFQAVQQLYNVRARVFWIHNTGPIGCLPYSYIYYE 174
Query: 228 HKNEKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNA 287
K KGN+DANGCVKP N+LAQE+NRQLKDQVFQLRR FPLAKFTYVDVYT KY LI+N
Sbjct: 175 PK--KGNIDANGCVKPQNDLAQEFNRQLKDQVFQLRRKFPLAKFTYVDVYTAKYELINNT 348
Query: 288 RNQGFVNPLEFCCGSYQGNEI 308
RNQGFV+PLEFCCGSY G I
Sbjct: 349 RNQGFVSPLEFCCGSYYGYHI 411
>TC205001 similar to UP|O82681 (O82681) Lanatoside 15'-O-acetylesterase
precursor, partial (62%)
Length = 1384
Score = 184 bits (466), Expect = 7e-47
Identities = 111/317 (35%), Positives = 161/317 (50%), Gaps = 7/317 (2%)
Frame = +1
Query: 24 CVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFISEELELPY 83
C + AI+NFGDSNSDTG + + A P G+++F GR DGRLI+DF+++ L LPY
Sbjct: 151 CDFEAIFNFGDSNSDTGGFHTSFPAQPAPYGMTYFKKPVGRASDGRLIVDFLAQGLGLPY 330
Query: 84 LSSYLNSVGSNYRHGANFAVASAPIRP-----IIAGLT--YLGFQVSQFILFKSHTKILF 136
LS YL S+GS+Y HGANFA +++ + P ++GL+ L Q+ Q FK+
Sbjct: 331 LSPYLQSIGSDYTHGANFASSASTVIPPTTSFSVSGLSPFSLSVQLRQMEQFKAKVDEFH 510
Query: 137 DQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQA 196
T + +P + F KA+YT IGQND + + VR S+P I+SQ A
Sbjct: 511 QTGTRISSGTKIPSPDIFGKALYTFYIGQNDFTSKI-AATGGIDGVRGSLPHIVSQINAA 687
Query: 197 VQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLK 256
+++LY + R F + N GP+ C P Y PH + D GC+ HN +YN+ L+
Sbjct: 688 IKELYAQGGRAFMVFNLGPVGCYPGYLVELPHAT--SDYDEFGCIVSHNNAVNDYNKLLR 861
Query: 257 DQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGSYQGNEIHYCGKKSI 316
D + Q A Y D ++ L + G CCG Y G ++ K I
Sbjct: 862 DTLTQTGESLVDASLIYADTHSALLELFHHPTFYGLKYNTRTCCG-YGGGVYNFNPK--I 1032
Query: 317 KNGTVYGIACDDSSTYI 333
G + ACD+ Y+
Sbjct: 1033LCGHMLASACDEPQNYV 1083
>BE329784
Length = 454
Score = 157 bits (396), Expect(2) = 7e-43
Identities = 74/96 (77%), Positives = 81/96 (84%)
Frame = +1
Query: 196 AVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQL 255
A Q+LYN ARVFWIHNTGPI C+PY Y +Y K KGN+DANGCVKP N+LAQE+NRQL
Sbjct: 1 AFQQLYNVGARVFWIHNTGPIGCLPYSYIYYEPK--KGNIDANGCVKPQNDLAQEFNRQL 174
Query: 256 KDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQG 291
KDQVFQLRR FPLAKFTYVDVYT KY LI+N RNQG
Sbjct: 175 KDQVFQLRRKFPLAKFTYVDVYTAKYELINNTRNQG 282
Score = 35.0 bits (79), Expect(2) = 7e-43
Identities = 13/15 (86%), Positives = 14/15 (92%)
Frame = +2
Query: 291 GFVNPLEFCCGSYQG 305
GFV+PLEFCCGSY G
Sbjct: 410 GFVSPLEFCCGSYYG 454
>TC229368 similar to PIR|T48618|T48618 early nodule-specific protein-like -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(61%)
Length = 1238
Score = 164 bits (415), Expect = 5e-41
Identities = 84/170 (49%), Positives = 112/170 (65%), Gaps = 7/170 (4%)
Frame = +2
Query: 53 NGISFFGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPII 112
+G FF SGR CDGRLI+DFI+E+L LPYLS+YLNS+G+NYRHGANFA + IR
Sbjct: 2 DGEGFFHKPSGRDCDGRLIVDFIAEKLNLPYLSAYLNSLGTNYRHGANFATGGSTIRKQN 181
Query: 113 AGLTYLG-------FQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQ 165
+ G Q+ QF FK+ TK L+++ P +S +P E+FSKA+YT DIGQ
Sbjct: 182 ETIFQYGISPFSLDIQIVQFNQFKARTKQLYEEAKAPHEKSKLPVPEEFSKALYTFDIGQ 361
Query: 166 NDIGYGLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGP 215
ND+ G +K N +++R S+PDIL+Q AV+ +Y + R FWIHNT P
Sbjct: 362 NDLSVGFRKMNF--DQIRESMPDILNQLANAVKNIYQQGGRYFWIHNTSP 505
Score = 117 bits (292), Expect = 1e-26
Identities = 63/138 (45%), Positives = 82/138 (58%), Gaps = 3/138 (2%)
Frame = +3
Query: 199 KLY-NEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLKD 257
K+Y N+E P C+P FY H +G LD GCVK N +A E+N+QLKD
Sbjct: 453 KIYTNKEGDTSGYTTLAPFGCMPVQ-LFYKHNIPEGYLDQYGCVKDQNVMATEFNKQLKD 629
Query: 258 QVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGSYQGNEIH-YCGKKSI 316
+V +LR P A TYVDVY KY LISN + +GFV+P++ CCG Y N+ H +CG
Sbjct: 630 RVIKLRTELPEAAITYVDVYAAKYALISNTKKEGFVDPMKICCG-YHVNDTHIWCGNLGT 806
Query: 317 KNG-TVYGIACDDSSTYI 333
NG V+G AC++ S YI
Sbjct: 807 DNGKDVFGSACENPSQYI 860
>TC230338 similar to UP|Q8W0Y6 (Q8W0Y6) Enod8.2, partial (49%)
Length = 737
Score = 163 bits (412), Expect = 1e-40
Identities = 92/212 (43%), Positives = 131/212 (61%), Gaps = 5/212 (2%)
Frame = +2
Query: 14 NLCVACPSKKCVYPAIYNFGDSNSDTGAGYAT--MAAVEHPNGISFFGSISGRCCDGRLI 71
N +A C +PAI+NFG SN+DTG A+ +AA + PNG ++F +GR DGRLI
Sbjct: 110 NPAMATKQYYCDFPAIFNFGASNADTGGLAASFFVAAPKSPNGETYFHRPAGRFSDGRLI 289
Query: 72 LDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRP---IIAGLTYLGFQVSQFILF 128
+DF+++ LPYLS YL+S+G+N+ GA+FA A + I P + LG Q SQF F
Sbjct: 290 IDFLAQSFGLPYLSPYLDSLGTNFSRGASFATAGSTIIPQQSFRSSPFSLGVQYSQFQRF 469
Query: 129 KSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPD 188
K T+ + +Q + +P+ E F +A+YT DIGQND+ G N + ++ +IPD
Sbjct: 470 KPTTQFIREQ--GGVFATLMPKEEYFHEALYTFDIGQNDLTAGF-FGNMTLQQFNATIPD 640
Query: 189 ILSQFTQAVQKLYNEEARVFWIHNTGPIECIP 220
I+ FT ++ +YN AR FWIHNTGPI C+P
Sbjct: 641 IIKSFTSNIKNIYNMGARSFWIHNTGPIGCLP 736
>TC223306 weakly similar to PIR|T48618|T48618 early nodule-specific
protein-like - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (25%)
Length = 669
Score = 129 bits (325), Expect = 1e-30
Identities = 66/141 (46%), Positives = 86/141 (60%), Gaps = 4/141 (2%)
Frame = +1
Query: 197 VQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEK---GNLDANGCVKPHNELAQEYNR 253
VQ L AR FWIHNTGPI C+P + N G LD NGC+ N++A+E+N+
Sbjct: 16 VQTLLGLGARTFWIHNTGPIGCLPVAMPVHNAMNTTPGAGYLDQNGCINYQNDMAREFNK 195
Query: 254 QLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGSYQGNEIHYCGK 313
+LK+ V +LR FP A YVD+++ KY LISNA +GFV+P CCG +Q YCG
Sbjct: 196 KLKNTVVKLRVQFPDASLIYVDMFSAKYELISNANKEGFVDPSGICCGYHQDGYHLYCGN 375
Query: 314 KSIKNG-TVYGIACDDSSTYI 333
K+I NG ++ CDD S YI
Sbjct: 376 KAIINGKEIFADTCDDPSKYI 438
>TC217532 weakly similar to UP|Q9LHW8 (Q9LHW8) ESTs AU075812(E60850), partial
(54%)
Length = 1372
Score = 125 bits (313), Expect = 4e-29
Identities = 101/346 (29%), Positives = 156/346 (44%), Gaps = 16/346 (4%)
Frame = +1
Query: 4 MTLIYILCFFNLCVACPSKKCVYPAIYNFGDSNSDTGAGY-----ATMAAVEHPNGISFF 58
+ L+ +L AC Y ++++FGDS +DTG Y + + P G + F
Sbjct: 64 IVLVLLLLLAATVTAC------YTSLFSFGDSLTDTGNLYFISPRQSPDCLLPPYGQTHF 225
Query: 59 GSISGRCCDGRLILDFISEELELPYLSSYLNSVG-----SNYRHGANFAVASAPIRPIIA 113
+GRC DGRLILDF++E L LPY+ YL N G NFAVA A
Sbjct: 226 HRPNGRCSDGRLILDFLAESLGLPYVKPYLGFKNGAVKRGNIEQGVNFAVAGATALD-RG 402
Query: 114 GLTYLGFQVSQFILFKSHTKILFDQRTEPPL-RSGVPRTEDFSKAIYTI-DIGQNDIGYG 171
GF V F ++ + + P L S + +++ + +IG ND GY
Sbjct: 403 FFEEKGFAVDVTANFSLGVQLDWFKELLPSLCNSSSSCKKVIGSSLFIVGEIGGNDYGYP 582
Query: 172 LQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNE 231
L + + + V IP ++S T A+++L + A F + + P+ C P Y + +
Sbjct: 583 LSETTAFGDLV-TYIPQVISVITSAIRELIDLGAVTFMVPGSLPLGCNPAYLTIFA-TID 756
Query: 232 KGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQG 291
K D GC+K N + +N L+ ++ +LR ++PL Y D + ++ G
Sbjct: 757 KEEYDQAGCLKWLNTFYEYHNELLQIEINRLRVLYPLTNIIYADYFNAALEFYNSPEQFG 936
Query: 292 F-VNPLEFCCGS---YQGNEIHYCGKKSIKNGTVYGIACDDSSTYI 333
F N L+ CCG Y NE CG + +ACDD S Y+
Sbjct: 937 FGGNVLKVCCGGGGPYNYNETAMCGDAGV-------VACDDPSQYV 1053
>TC220456 weakly similar to UP|O81262 (O81262) Early nodule-specific protein,
partial (43%)
Length = 587
Score = 125 bits (313), Expect = 4e-29
Identities = 73/155 (47%), Positives = 101/155 (65%), Gaps = 7/155 (4%)
Frame = +1
Query: 18 ACPSKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFISE 77
A +K+CV+PAI+NFGDSNSDTG A++ A P G ++F +GR DGRL++DFI++
Sbjct: 100 AFATKECVFPAIFNFGDSNSDTGGLAASLIAPTPPYGETYFHRPAGRFSDGRLVIDFIAK 279
Query: 78 ELELPYLSSYLNSVGSNYRHGANFAVASAPIR---PII--AGLT--YLGFQVSQFILFKS 130
LPYLS+YL+S+G+N+ HGANFA +++ IR II G + YL Q +QF KS
Sbjct: 280 SFGLPYLSAYLDSLGTNFSHGANFATSASTIRLPTSIIPQGGFSPFYLDIQYTQFRDSKS 459
Query: 131 HTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQ 165
T+ F + S +P+ E F KA+YT DIGQ
Sbjct: 460 RTQ--FIRHQGGVFASLMPKEEYFDKALYTFDIGQ 558
>TC232240 weakly similar to UP|Q9FXE5 (Q9FXE5) F12A21.4, partial (38%)
Length = 469
Score = 113 bits (283), Expect = 1e-25
Identities = 63/149 (42%), Positives = 90/149 (60%), Gaps = 7/149 (4%)
Frame = +2
Query: 17 VACPSKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFIS 76
V+ +C++PAI+N GDSNSDTG A PNGI++F S +GR DGRLI+DFI+
Sbjct: 29 VSGSESECIFPAIFNLGDSNSDTGGLSAAFGQAPPPNGITYFHSPNGRFSDGRLIIDFIA 208
Query: 77 EELELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLGF-------QVSQFILFK 129
E L YL +YL+SV SN+ HGANFA A + +RP ++ G+ Q QF FK
Sbjct: 209 ESSGLAYLRAYLDSVASNFTHGANFATAGSTVRPQNTTISQSGYSPISLDVQFVQFSDFK 388
Query: 130 SHTKILFDQRTEPPLRSGVPRTEDFSKAI 158
+ +K++ Q + +P+ E FS+A+
Sbjct: 389 TRSKLVRQQ--GGVFKELLPKEEYFSQAL 469
>TC233041 weakly similar to UP|Q8W0Y7 (Q8W0Y7) Enod8.3 (Fragment), partial
(35%)
Length = 748
Score = 74.7 bits (182), Expect(2) = 1e-25
Identities = 35/78 (44%), Positives = 49/78 (61%)
Frame = +2
Query: 182 VRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCV 241
V+ I D+L Q + ++ +Y E FWIHNTGP+ C+PY YP K + +D GC
Sbjct: 20 VKAYIXDVLGQXSNVIKGVYGEGGXSFWIHNTGPLGCLPYMLDRYPMKPTQ--MDEFGCA 193
Query: 242 KPHNELAQEYNRQLKDQV 259
KP NE+AQ +NR+LK+ V
Sbjct: 194 KPFNEVAQYFNRKLKEVV 247
Score = 59.7 bits (143), Expect(2) = 1e-25
Identities = 37/77 (48%), Positives = 40/77 (51%), Gaps = 5/77 (6%)
Frame = +3
Query: 262 LRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCC---GSYQGNEIHYCGKKSIKN 318
LR+ P A TYVDVYTVKYTLIS+A GF + CC G Y N CG N
Sbjct: 255 LRKELPGAAITYVDVYTVKYTLISHAHKYGFEQGVIACCGHGGKYNFNNTERCGATKRVN 434
Query: 319 GTVYGIA--CDDSSTYI 333
GT IA C D S I
Sbjct: 435 GTEIVIANSCKDLSVRI 485
>TC209518 weakly similar to UP|O82076 (O82076) Lipase homolog (Fragment),
partial (56%)
Length = 874
Score = 112 bits (280), Expect = 2e-25
Identities = 63/174 (36%), Positives = 93/174 (53%), Gaps = 5/174 (2%)
Frame = +1
Query: 165 QNDIGYGLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYF 224
QND+ K N S +V + IP ++++ AV+ LYN+ AR FW+HNTGP+ C+P
Sbjct: 1 QNDLADSFAK-NLSYAQVIKKIPAVITEIENAVKNLYNDGARKFWVHNTGPLGCLPKVLA 177
Query: 225 FYPHKNEKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLI 284
+K +LD+ GC+ +N A+ +N L +LR F A YVD+Y +KY LI
Sbjct: 178 LA----QKKDLDSLGCLSSYNSAARLFNEALLHSSQKLRSEFKDATLVYVDIYAIKYDLI 345
Query: 285 SNARNQGFVNPLEFCCGSYQGNEIHY-----CGKKSIKNGTVYGIACDDSSTYI 333
+NA GF NPL CCG Y G ++ CG+ + CD+ + Y+
Sbjct: 346 TNAAKYGFSNPLMVCCG-YGGPPYNFDVRVTCGQPGYQ-------VCDEGARYV 483
>TC207591 weakly similar to UP|Q9LHW8 (Q9LHW8) ESTs AU075812(E60850), partial
(57%)
Length = 1290
Score = 97.4 bits (241), Expect = 8e-21
Identities = 84/278 (30%), Positives = 130/278 (46%), Gaps = 12/278 (4%)
Frame = +1
Query: 67 DGRLILDFISEELELPYLSSYLN-SVGSNYRHGANFAVASAPIRP----IIAGLT-YLGF 120
DGRL++DFI+E +LPYL YL + + + G NFAVA A I AGL YL
Sbjct: 7 DGRLMIDFIAEAYDLPYLPPYLALTKDKDIQRGVNFAVAGATALDAKFFIEAGLAKYLWT 186
Query: 121 QVSQFILFKSHTKILFDQRTEPPLRSGVPRTED-FSKAIYTI-DIGQNDIGYGLQKPNSS 178
S I ++ + ++ +P L + + F ++++ + +IG ND Y N +
Sbjct: 187 NNSLNI------QLGWFKKLKPSLCTTKQDCDSYFKRSLFLVGEIGGNDYNYAAIAGNIT 348
Query: 179 EEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDAN 238
+ ++ ++P ++ T A+ +L E AR + PI C Y + +N K + D +
Sbjct: 349 Q--LQATVPPVVEAITAAINELIAEGARELLVPGNFPIGCSALYLTLFRSEN-KEDYDES 519
Query: 239 GCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVN-PLE 297
GC+K N A+ +N++LK + LR+ P A+ Y D Y + GF N L
Sbjct: 520 GCLKTFNGFAEYHNKELKLALETLRKKNPHARILYADYYGAAKRFFHAPGHHGFTNGALR 699
Query: 298 FCCGS---YQGNEIHYCGKKSIKNGTVYGIACDDSSTY 332
CCG + N CG K AC D STY
Sbjct: 700 ACCGGGGPFNFNISARCGHTGSK-------ACADPSTY 792
>AW830391
Length = 419
Score = 95.5 bits (236), Expect = 3e-20
Identities = 53/116 (45%), Positives = 76/116 (64%), Gaps = 3/116 (2%)
Frame = +3
Query: 1 MNTMTLIYILCFFNLCVACP-SKKCVYPAIYNFGDSNSDTGAGYATMA-AVEHPNGISFF 58
M + +I F++C+A S + YPA++NFGDS SDTG A + V PNG +F
Sbjct: 54 MASKNVILQFVLFSMCLAMANSVEFKYPAVFNFGDSYSDTGELAAGLGFQVAPPNGQDYF 233
Query: 59 GSISGRCCDGRLILDFISEELELPYLSSYLNSVG-SNYRHGANFAVASAPIRPIIA 113
SGR CDGRLI+DF+ + ++LP+L++YL+S+G N+R G+NFA A+A I P A
Sbjct: 234 KIPSGRFCDGRLIVDFLMDAMDLPFLNAYLDSLGLPNFRKGSNFAAAAATILPATA 401
>TC212934 similar to UP|O82681 (O82681) Lanatoside 15'-O-acetylesterase
precursor, partial (30%)
Length = 442
Score = 62.4 bits (150), Expect(2) = 1e-19
Identities = 31/68 (45%), Positives = 41/68 (59%)
Frame = +2
Query: 7 IYILCFFNLCVACPSKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCC 66
I +LC F+L +C + AI+NFGDSNSDTG YA P G+++F +GR
Sbjct: 44 IVLLCLFSLS----HSECNFKAIFNFGDSNSDTGGFYAAFPGESGPYGMTYFKKPAGRAS 211
Query: 67 DGRLILDF 74
DGRLI+ F
Sbjct: 212 DGRLIIGF 235
Score = 51.6 bits (122), Expect(2) = 1e-19
Identities = 28/67 (41%), Positives = 45/67 (66%), Gaps = 7/67 (10%)
Frame = +3
Query: 71 ILDFISEELELPYLSSYLNSVGSNYRHGANFA-VASAPIRP----IIAGLT--YLGFQVS 123
+LDF+++ L LP+LS YL S+GS+Y+HGAN+A +AS + P + G++ L Q++
Sbjct: 225 LLDFLAQALGLPFLSPYLQSIGSDYKHGANYATMASTVLMPNTSLFVTGISPFSLAIQLN 404
Query: 124 QFILFKS 130
Q FK+
Sbjct: 405 QMKQFKT 425
>TC207761 weakly similar to UP|Q8L8G1 (Q8L8G1) Lipase SIL1, partial (46%)
Length = 1609
Score = 68.2 bits (165), Expect(2) = 2e-18
Identities = 52/212 (24%), Positives = 95/212 (44%), Gaps = 1/212 (0%)
Frame = +1
Query: 123 SQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEV 182
+Q FK +KIL + + T +KA+Y I+IG ND L + NSS
Sbjct: 724 TQLSYFKKVSKILSQELGD------AETTTLLAKAVYLINIGSNDYLVSLTE-NSSVFTA 882
Query: 183 RRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVK 242
+ + ++ T ++ ++ R F + N + CIP N CV+
Sbjct: 883 EKYVDMVVGNLTTVIKGIHKTGGRKFGVLNQSALGCIPLVKALL-------NGSKGSCVE 1041
Query: 243 PHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGS 302
+ LA+ +N L ++ +L++ K++YVD + + + L++N G CCGS
Sbjct: 1042EASALAKLHNGVLSVELEKLKKQLEGFKYSYVDFFNLSFDLMNNPSKYGLKEGGMACCGS 1221
Query: 303 YQGNEIHYC-GKKSIKNGTVYGIACDDSSTYI 333
+ C GK+++K+ + C++ S Y+
Sbjct: 1222GPYRRYYSCGGKRAVKDYEL----CENPSDYV 1305
Score = 42.0 bits (97), Expect(2) = 2e-18
Identities = 24/55 (43%), Positives = 29/55 (52%)
Frame = +3
Query: 52 PNGISFFGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASA 106
P G +FF SGR DGR+I D I++ +LP YL Y G NFA A A
Sbjct: 507 PYGETFFNYPSGRFSDGRVIPDLIADYAKLPLSPPYLFPGYQRYLDGVNFASAGA 671
>TC231116 similar to UP|Q9FFC6 (Q9FFC6) GDSL-motif lipase/hydrolase-like
protein, partial (94%)
Length = 1541
Score = 87.0 bits (214), Expect = 1e-17
Identities = 89/317 (28%), Positives = 131/317 (41%), Gaps = 10/317 (3%)
Frame = +1
Query: 27 PAIYNFGDSNSDTGAGYATMAAVEH---PNGISFFGSI-SGRCCDGRLILDFISEELELP 82
PAI+ FGDS D G + V+ P G F +GR C+G+L DFI++ L
Sbjct: 442 PAIFTFGDSIVDVGNNNHQLTIVKANFPPYGRDFENHFPTGRFCNGKLATDFIADILGFT 621
Query: 83 -YLSSYLN--SVGSNYRHGANFAVASAPIRPIIAGLTYLGFQVSQFILFKSHTKILFDQR 139
Y +YLN + G N +GANFA AS+ + + L Y +S+ + + + +
Sbjct: 622 SYQPAYLNLKTKGKNLLNGANFASASSGYFELTSKL-YSSIPLSKQLEYYKECQTKLVEA 798
Query: 140 TEPPLRSGVPRTEDFSKAIYTIDIGQNDI--GYGLQKPNSSEEEVRRSIPDILSQFTQAV 197
S + S AIY I G +D Y + + + +L ++ +
Sbjct: 799 AGQSSASSI-----ISDAIYLISAGTSDFVQNYYINPLLNKLYTTDQFSDTLLRCYSNFI 963
Query: 198 QKLYNEEARVFWIHNTGPIECIPYYY-FFYPHKNEKGNLDANGCVKPHNELAQEYNRQLK 256
Q LY AR + + PI C+P F H NE CV N A +N +L
Sbjct: 964 QSLYALGARRIGVTSLPPIGCLPAVITLFGAHINE--------CVTSLNSDAINFNEKLN 1119
Query: 257 DQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGSYQGNEIHYCGKKSI 316
L+ M P D+Y Y L + GF + CCG+ C KKSI
Sbjct: 1120TTSQNLKNMLPGLNLVVFDIYQPLYDLATKPSENGFFEARKACCGTGLIEVSILCNKKSI 1299
Query: 317 KNGTVYGIACDDSSTYI 333
GT C ++S Y+
Sbjct: 1300--GT-----CANASEYV 1329
>CD409722
Length = 605
Score = 81.3 bits (199), Expect = 6e-16
Identities = 43/109 (39%), Positives = 63/109 (57%), Gaps = 6/109 (5%)
Frame = -2
Query: 231 EKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQ 290
+ LD GCV HN+ A+ +N QL+ +L+ +P + TYVD++T+K +LI+N
Sbjct: 568 DSSKLDGLGCVSSHNQAAKTFNLQLRALCTKLQGQYPDSNVTYVDIFTIKSSLIANYSRY 389
Query: 291 GFVNPLEFCCGSYQGNEIHY-----CGKKSIKNG-TVYGIACDDSSTYI 333
GF P+ CCG Y G ++Y CG+ NG T+ AC+DSS YI
Sbjct: 388 GFEQPIMACCG-YGGPPLNYDSRVSCGETKTFNGTTITAKACNDSSEYI 245
>TC232569
Length = 974
Score = 77.4 bits (189), Expect = 9e-15
Identities = 75/259 (28%), Positives = 115/259 (43%), Gaps = 10/259 (3%)
Frame = +2
Query: 27 PAIYNFGDSNSDTGAG---YATMAAVEHPNGISFFGSISGRCCDGRLILDFISEELELPY 83
P + GDS SD G ++ P GI + +GR +G+ I+DFISE L
Sbjct: 191 PCTFVLGDSLSDNGNNNNLQTNASSNYRPYGIDYPTGPTGRFTNGKNIIDFISEYLGFTE 370
Query: 84 -LSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLGFQVSQFILFKSHTKILFDQRTEP 142
+ N+ GS+ GAN+A +A I P +G +LG + ++H + +
Sbjct: 371 PIPPNANTSGSDILKGANYASGAAGI-PFKSG-KHLGDNIHLGEQIRNHRATI--TKIVR 538
Query: 143 PLRSGVPRTEDFSKAIYTIDIGQNDI--GYGLQK--PNSSEEEVRRSIPDILSQFTQAVQ 198
L E K +Y ++IG ND Y L + P S + R ++ Q++ ++
Sbjct: 539 RLGGSGRAREYLKKCLYYVNIGSNDYINNYFLPQFYPTSRTYTLERYTDILIKQYSDDIK 718
Query: 199 KLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANG--CVKPHNELAQEYNRQLK 256
KL+ AR F I G I C P N NG CV N A ++ +LK
Sbjct: 719 KLHRSGARKFAIVGLGLIGCTP---------NAISRRGTNGEVCVAELNNAAFLFSNKLK 871
Query: 257 DQVFQLRRMFPLAKFTYVD 275
QV Q + FP +KF++V+
Sbjct: 872 SQVDQFKNTFPDSKFSFVN 928
>TC217084
Length = 1310
Score = 76.3 bits (186), Expect = 2e-14
Identities = 80/297 (26%), Positives = 112/297 (36%), Gaps = 13/297 (4%)
Frame = +3
Query: 28 AIYNFGDSNSDTGAGY---ATMAAVEHPNGI-SFFGSISGRCCDGRLILDFISEEL---- 79
A + FGDS D G T A +P GI S SGR +G + D ISE++
Sbjct: 165 AFFVFGDSLVDNGNNNFLATTARADSYPYGIDSASHRASGRFSNGLNMPDLISEKIGSEP 344
Query: 80 ELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLGFQVSQFILFKSHTKILFDQR 139
LPYLS LN G GANFA A I L G Q I +
Sbjct: 345 TLPYLSPQLN--GERLLVGANFASAGIGI------LNDTGIQFINIIRITEQLAYFKQYQ 500
Query: 140 TEPPLRSGVPRTEDF-SKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPD----ILSQFT 194
G +T + +KA+ I +G ND S ++PD ++S++
Sbjct: 501 QRVSALIGEEQTRNLVNKALVLITLGGNDFVNNYYLVPFSARSREYALPDYVVFLISEYR 680
Query: 195 QAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQ 254
+ + LY AR + TGP+ C+P + E C +N Q
Sbjct: 681 KILANLYELGARRVLVTGTGPLGCVPAELAMHSQNGE--------CATELQRAVNLFNPQ 836
Query: 255 LKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGSYQGNEIHYC 311
L + +L F + +T+ +SN + GFV CCG N I C
Sbjct: 837 LVQLLHELNTQIGSDVFISANAFTMHLDFVSNPQAYGFVTSKVACCGQGAYNGIGLC 1007
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.139 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,706,146
Number of Sequences: 63676
Number of extensions: 241176
Number of successful extensions: 1447
Number of sequences better than 10.0: 120
Number of HSP's better than 10.0 without gapping: 1378
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1387
length of query: 341
length of database: 12,639,632
effective HSP length: 98
effective length of query: 243
effective length of database: 6,399,384
effective search space: 1555050312
effective search space used: 1555050312
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC136841.9


