

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.7 + phase: 0 /pseudo
(381 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
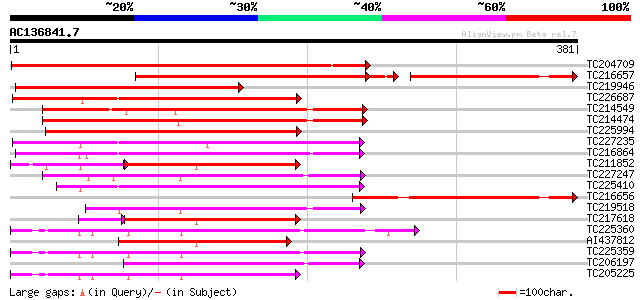
Score E
Sequences producing significant alignments: (bits) Value
TC204709 similar to UP|Q8H0X4 (Q8H0X4) Serine/threonine-protein ... 303 6e-83
TC216657 similar to UP|MHK_ARATH (P43294) Serine/threonine-prote... 292 2e-79
TC219946 similar to UP|O49669 (O49669) Kinase-like protein (Frag... 222 2e-58
TC226687 homologue to UP|MMK2_MEDSA (Q40353) Mitogen-activated p... 160 7e-40
TC214549 homologue to UP|O82135 (O82135) Cdc2, complete 158 3e-39
TC214474 homologue to UP|CDC2_VIGAC (Q41639) Cell division contr... 156 2e-38
TC225994 similar to UP|Q7XUF4 (Q7XUF4) OJ991113_30.14 protein, p... 153 1e-37
TC227235 homologue to UP|Q6TAR9 (Q6TAR9) Mitogen activated prote... 151 4e-37
TC216864 homologue to UP|Q9M6R8 (Q9M6R8) MAP kinase PsMAPK2, com... 149 3e-36
TC211852 homologue to UP|Q9LDC1 (Q9LDC1) CRK1 protein, partial (... 116 2e-33
TC227247 UP|Q6T2Z8 (Q6T2Z8) Cyclin-dependent kinases CDKB, complete 139 2e-33
TC225410 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, partial (... 139 2e-33
TC216656 weakly similar to UP|MHK_ARATH (P43294) Serine/threonin... 134 7e-32
TC219518 similar to UP|Q9FYT8 (Q9FYT8) Cyclin-dependent kinase B... 125 2e-29
TC217618 similar to GB|AAO64868.1|29028978|BT005933 At5g50860 {A... 121 3e-28
TC225360 homologue to UP|MSK1_MEDSA (P51137) Glycogen synthase k... 122 3e-28
AI437812 122 3e-28
TC225359 homologue to UP|MSK1_MEDSA (P51137) Glycogen synthase k... 121 6e-28
TC206197 homologue to UP|MAPK_PEA (Q06060) Mitogen-activated pro... 118 4e-27
TC205225 homologue to UP|KSG1_ARATH (P43288) Shaggy-related prot... 118 4e-27
>TC204709 similar to UP|Q8H0X4 (Q8H0X4) Serine/threonine-protein kinase Mak
(Male germ cell-associated kinase)-like protein, partial
(59%)
Length = 2140
Score = 303 bits (777), Expect = 6e-83
Identities = 146/241 (60%), Positives = 195/241 (80%)
Frame = +2
Query: 2 RTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDC 61
+T E+VA+K++K+K+ WEE NLRE+K+LRKMNH NI+KL+EV+RE++ L+F+FEYM+C
Sbjct: 602 QTGEVVAIKKMKKKYYSWEECVNLREVKSLRKMNHPNIVKLKEVIRESDILYFVFEYMEC 781
Query: 62 NLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFG 121
NLYQL+K+REK FSE E+R + Q+ QGL++MH++G+FHRDLKPENLLVT D +KIADFG
Sbjct: 782 NLYQLMKDREKLFSEAEVRNWCFQVFQGLAYMHQRGYFHRDLKPENLLVTKDFIKIADFG 961
Query: 122 LAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESE 181
LARE+SS PPYT+YVSTRWYRAPEVLLQS YT VDMWA+GAI+AELF+L P+FPG SE
Sbjct: 962 LAREISSQPPYTEYVSTRWYRAPEVLLQSYMYTSKVDMWAMGAIMAELFSLRPLFPGASE 1141
Query: 182 IDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITAR 241
D++YKI ++G P + G +R +++ + +A V LS +IP+AS +AI LIT+
Sbjct: 1142ADEIYKICGVIGNPTFESWADGLKLARDINYQFPQ-LAGVHLSALIPSASDDAISLITSL 1318
Query: 242 C 242
C
Sbjct: 1319C 1321
>TC216657 similar to UP|MHK_ARATH (P43294) Serine/threonine-protein kinase
MHK , partial (41%)
Length = 1393
Score = 292 bits (747), Expect = 2e-79
Identities = 141/158 (89%), Positives = 150/158 (94%)
Frame = +2
Query: 85 QMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFGLAREVSSMPPYTQYVSTRWYRAP 144
Q+LQGLSHMHKKGFFHRDLKPEN+LVTNDVLKIADFGLAREVSSMPPYTQYVSTRWYRAP
Sbjct: 8 QVLQGLSHMHKKGFFHRDLKPENMLVTNDVLKIADFGLAREVSSMPPYTQYVSTRWYRAP 187
Query: 145 EVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILGMPDSTCFTIGA 204
EVLL++PCYTPAVDMWA+GAILAELFTLTPIFPGESEIDQ+YKIY ILGMPDST FTIG
Sbjct: 188 EVLLRAPCYTPAVDMWAVGAILAELFTLTPIFPGESEIDQLYKIYGILGMPDSTAFTIGE 367
Query: 205 NNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITARC 242
NNS+LLD V HEVV PVKLS+IIPNAS+EAIDLIT C
Sbjct: 368 NNSQLLDIVAHEVVPPVKLSNIIPNASLEAIDLITQCC 481
Score = 125 bits (315), Expect(2) = 3e-30
Identities = 73/113 (64%), Positives = 79/113 (69%), Gaps = 1/113 (0%)
Frame = +1
Query: 270 VNTRVPRSLSDPLELKLSNKRVKPNLELKLHDFGPDPDDCFLGLTLAVKPSVSNLDVVQN 329
V+ VP SDPLELKLS+KR KPNLELKL DFGPDPDDCFLGLTL VKPSVSN DVVQN
Sbjct: 658 VDAWVPCPPSDPLELKLSSKRAKPNLELKLQDFGPDPDDCFLGLTLGVKPSVSNSDVVQN 837
Query: 330 ARQGMGEN-LV*LDAL*KCRICYFARILMTIQTNQPDQNGVHSSAETSLSLSF 381
QG+ EN L D F +L PDQNG+H+SAETSLSLSF
Sbjct: 838 VSQGVRENVLFCSDFNDHSDQSVFWTLL------SPDQNGIHNSAETSLSLSF 978
Score = 23.9 bits (50), Expect(2) = 3e-30
Identities = 12/22 (54%), Positives = 14/22 (63%)
Frame = +3
Query: 240 ARCGSVTATSFFPSCSILHKVS 261
ARC SV ATS FP C + +S
Sbjct: 618 ARCRSVIATSIFP-CGCMGSLS 680
>TC219946 similar to UP|O49669 (O49669) Kinase-like protein (Fragment),
partial (61%)
Length = 806
Score = 222 bits (566), Expect = 2e-58
Identities = 103/153 (67%), Positives = 130/153 (84%)
Frame = +1
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E+VA+K++K+K+ WEE NLRE+K+LRKMNH NI+KL+EV+RE + L +FEYM+ NLY
Sbjct: 346 EVVAIKKMKKKYYSWEECVNLREVKSLRKMNHANIVKLKEVIRECDTLCLVFEYMEYNLY 525
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFGLAR 124
QL+K REK FSE E+R + Q+ QGL++MH++G+FHRDLKPENLLVT DV+KIADFGLAR
Sbjct: 526 QLMKNREKLFSENEVRNWCFQVFQGLAYMHQRGYFHRDLKPENLLVTKDVIKIADFGLAR 705
Query: 125 EVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAV 157
E+SS PPYT+YVSTRWYRAPEVLLQS Y+ V
Sbjct: 706 EISSQPPYTEYVSTRWYRAPEVLLQSHLYSSKV 804
>TC226687 homologue to UP|MMK2_MEDSA (Q40353) Mitogen-activated protein
kinase homolog MMK2 , partial (97%)
Length = 1783
Score = 160 bits (406), Expect = 7e-40
Identities = 87/201 (43%), Positives = 130/201 (64%), Gaps = 7/201 (3%)
Frame = +3
Query: 3 TFEIVAVKRLKRKFCFW-EEYTNLREIKALRKMNHQNIIKLREVVR----EN-NELFFIF 56
T E VA+K++ F + LREIK LR M+H NI+ +++++R EN N+++ +
Sbjct: 408 TGEEVAIKKIGNAFDNRIDAKRTLREIKLLRHMDHANIMSIKDIIRPPQKENFNDVYLVS 587
Query: 57 EYMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVT-NDVL 115
E MD +L+Q+I+ ++ +++ R F+ Q+L+GL ++H HRDLKP NLL+ N L
Sbjct: 588 ELMDTDLHQIIRSNQQ-LTDDHCRYFLYQLLRGLKYVHSANVLHRDLKPSNLLLNANCDL 764
Query: 116 KIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPI 175
KIADFGLAR S T+YV TRWYRAPE+LL YT A+D+W++G IL E+ T P+
Sbjct: 765 KIADFGLARTTSETDFMTEYVVTRWYRAPELLLNCSEYTAAIDIWSVGCILGEIITRQPL 944
Query: 176 FPGESEIDQMYKIYCILGMPD 196
FPG+ + Q+ I ++G PD
Sbjct: 945 FPGKDYVHQLRLITELIGSPD 1007
>TC214549 homologue to UP|O82135 (O82135) Cdc2, complete
Length = 1216
Score = 158 bits (400), Expect = 3e-39
Identities = 84/224 (37%), Positives = 141/224 (62%), Gaps = 6/224 (2%)
Frame = +3
Query: 23 TNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLYQLIKEREKPFSEE--EIR 80
T +REI L++M H+NI++L++VV + L+ +FEY+D +L + + + F+++ +++
Sbjct: 294 TAIREISLLKEMQHRNIVRLQDVVHDEKSLYLVFEYLDLDLKKHM-DSSPEFAKDPRQVK 470
Query: 81 CFMKQMLQGLSHMHKKGFFHRDLKPENLLV--TNDVLKIADFGLAREVS-SMPPYTQYVS 137
F+ Q+L G+++ H HRDLKP+NLL+ + + LK+ADFGLAR + +T V
Sbjct: 471 MFLYQILCGIAYCHSHRVLHRDLKPQNLLIDRSTNALKLADFGLARAFGIPVRTFTHEVV 650
Query: 138 TRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILGMPDS 197
T WYRAPE+LL S Y+ VD+W++G I AE+ P+FPG+SEID+++KI+ I+G P+
Sbjct: 651 TLWYRAPEILLGSRQYSTPVDIWSVGCIFAEMVNQRPLFPGDSEIDELFKIFRIMGTPNE 830
Query: 198 TCFTIGANNSRLLDF-VGHEVVAPVKLSDIIPNASMEAIDLITA 240
+ + L DF P L +++PN +DL+++
Sbjct: 831 DTW---PGVTSLPDFKSAFPKWQPKDLKNVVPNLEPAGLDLLSS 953
>TC214474 homologue to UP|CDC2_VIGAC (Q41639) Cell division control protein 2
homolog (p34cdc2) , complete
Length = 1204
Score = 156 bits (394), Expect = 2e-38
Identities = 84/223 (37%), Positives = 138/223 (61%), Gaps = 5/223 (2%)
Frame = +2
Query: 23 TNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLYQLIKEREKPFSE-EEIRC 81
T +REI L++M H+NI++L++VV L+ +FEY+D +L + + + + +++
Sbjct: 218 TAIREISLLKEMQHRNIVRLQDVVHSEKRLYLVFEYLDLDLKKHMDSSPEFVKDPRQVKM 397
Query: 82 FMKQMLQGLSHMHKKGFFHRDLKPENLLVTN--DVLKIADFGLAREVS-SMPPYTQYVST 138
F+ Q+L G+++ H HRDLKP+NLL+ + LK+ADFGLAR + +T V T
Sbjct: 398 FLYQILCGIAYCHSHRVLHRDLKPQNLLIDRRTNSLKLADFGLARAFGIPVRTFTHEVVT 577
Query: 139 RWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILGMPDST 198
WYRAPE+LL S Y+ VD+W++G I AE+ P+FPG+SEID+++KI+ ILG P+
Sbjct: 578 LWYRAPEILLGSRHYSTPVDVWSVGCIFAEMVNRRPLFPGDSEIDELFKIFRILGTPNED 757
Query: 199 CFTIGANNSRLLDFVGHEVVAPVK-LSDIIPNASMEAIDLITA 240
+ + L DF P K L++++PN ++L+++
Sbjct: 758 TW---PGVTSLPDFKSTFPKWPSKDLANVVPNLDAAGLNLLSS 877
>TC225994 similar to UP|Q7XUF4 (Q7XUF4) OJ991113_30.14 protein, partial (43%)
Length = 2770
Score = 153 bits (386), Expect = 1e-37
Identities = 76/175 (43%), Positives = 120/175 (68%), Gaps = 3/175 (1%)
Frame = +2
Query: 25 LREIKALRKMNHQNIIKLREVVRENNE-LFFIFEYMDCNLYQLIKEREKPFSEEEIRCFM 83
LREI L NH +I+ ++EVV ++ + F + E+M+ +L L++ +++PFS EI+ +
Sbjct: 785 LREINILLSFNHPSIVNVKEVVVDDFDGTFMVMEHMEYDLKGLMEVKKQPFSMSEIKSLV 964
Query: 84 KQMLQGLSHMHKKGFFHRDLKPENLLVTND-VLKIADFGLAREVSS-MPPYTQYVSTRWY 141
+Q+L+G+ ++H HRDLK N+L+ +D LKI DFGL+R+ S + PYT V T WY
Sbjct: 965 RQLLEGVKYLHDNWVIHRDLKSSNILLNHDGELKICDFGLSRQYGSPLKPYTPVVVTLWY 1144
Query: 142 RAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILGMPD 196
RAPE+LL + Y+ ++DMW++G I+AEL P+F G+SE++Q+ KI+ LG PD
Sbjct: 1145RAPELLLGAKEYSTSIDMWSVGCIMAELIAKEPLFRGKSELEQLDKIFRTLGTPD 1309
>TC227235 homologue to UP|Q6TAR9 (Q6TAR9) Mitogen activated protein kinase 6,
partial (72%)
Length = 2041
Score = 151 bits (382), Expect = 4e-37
Identities = 93/248 (37%), Positives = 140/248 (55%), Gaps = 12/248 (4%)
Frame = +3
Query: 3 TFEIVAVKRLKRKFCFWEEYTN-LREIKALRKMNHQNIIKLREVV-----RENNELFFIF 56
T E VA+K++ F + T LREIK LR + H +I++++ ++ RE +++ +F
Sbjct: 33 TGEKVAIKKINDIFEHVSDATRILREIKLLRLLRHPDIVEIKHILLPPSRREFKDIYVVF 212
Query: 57 EYMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDV-L 115
E M+ +L+Q+IK + + E + F+ Q+L+G+ ++H FHRDLKP+N+L D L
Sbjct: 213 ELMESDLHQVIKANDD-LTPEHYQFFLYQLLRGMKYIHTANVFHRDLKPKNILANADCKL 389
Query: 116 KIADFGLAREVSSMPP----YTQYVSTRWYRAPEVLLQS-PCYTPAVDMWAIGAILAELF 170
KI DFGLAR + P +T YV+TRWYRAPE+ YTPA+D+W+IG I AEL
Sbjct: 390 KICDFGLARVAFNDTPTAIFWTDYVATRWYRAPELCGSFFSKYTPAIDIWSIGCIFAELL 569
Query: 171 TLTPIFPGESEIDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNA 230
T P+FPG++ + Q+ + +LG P N PV LS PNA
Sbjct: 570 TGKPLFPGKNVVHQLDLMTDLLGTPSLEAIARVRNEKARRYLSSMRKKKPVPLSQKFPNA 749
Query: 231 SMEAIDLI 238
A+ L+
Sbjct: 750 DPLALRLL 773
>TC216864 homologue to UP|Q9M6R8 (Q9M6R8) MAP kinase PsMAPK2, complete
Length = 1735
Score = 149 bits (375), Expect = 3e-36
Identities = 93/243 (38%), Positives = 142/243 (58%), Gaps = 9/243 (3%)
Frame = +2
Query: 5 EIVAVKRLKRKF-CFWEEYTNLREIKALRKMNHQNIIKLREV---VRENN--ELFFIFEY 58
E VA+K+++ F + LRE+K LR ++H+N+I L+++ V N+ +++ ++E
Sbjct: 464 EKVAIKKIQNAFENRVDALRTLRELKLLRHLHHENVIALKDIMMPVHRNSFKDVYLVYEL 643
Query: 59 MDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVT-NDVLKI 117
MD +L+Q+IK + S + + F+ Q+L+GL ++H HRDLKP NLL+ N LKI
Sbjct: 644 MDTDLHQIIKSSQA-LSNDHCQYFLFQLLRGLKYLHSANILHRDLKPGNLLINANCDLKI 820
Query: 118 ADFGLAREVSSMPPY-TQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIF 176
DFGLAR S + T+YV TRWYRAPE+LL Y ++D+W++G I AEL PIF
Sbjct: 821 CDFGLARTNCSKNQFMTEYVVTRWYRAPELLLCCDNYGTSIDVWSVGCIFAELLGRKPIF 1000
Query: 177 PGESEIDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAP-VKLSDIIPNASMEAI 235
PG ++Q+ I ILG +N + ++ +P LS + PNA AI
Sbjct: 1001PGSECLNQLKLIINILGSQREEDIEF-IDNPKAKKYIKSLPYSPGTPLSQLYPNAHPLAI 1177
Query: 236 DLI 238
DL+
Sbjct: 1178DLL 1186
>TC211852 homologue to UP|Q9LDC1 (Q9LDC1) CRK1 protein, partial (39%)
Length = 706
Score = 116 bits (291), Expect(2) = 2e-33
Identities = 56/122 (45%), Positives = 79/122 (63%), Gaps = 3/122 (2%)
Frame = +2
Query: 77 EEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTND-VLKIADFGLAR--EVSSMPPYT 133
++++C+M Q+L GL H H + HRD+K NLL+ N+ LKIADFGLA + + P T
Sbjct: 296 DQVKCYMHQLLSGLEHCHNRHVLHRDIKGSNLLIDNEGTLKIADFGLASIFDPNHKHPMT 475
Query: 134 QYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILG 193
V T WYR PE+LL + Y+ VD+W+ G IL EL PI PG +E++Q++KIY + G
Sbjct: 476 SRVVTLWYRPPELLLGATDYSVGVDLWSAGCILGELLAGKPIMPGRTEVEQLHKIYKLCG 655
Query: 194 MP 195
P
Sbjct: 656 SP 661
Score = 44.3 bits (103), Expect(2) = 2e-33
Identities = 29/85 (34%), Positives = 51/85 (59%), Gaps = 5/85 (5%)
Frame = +1
Query: 1 MRTFEIVAVKRLKRKFCFWEEYT---NLREIKALRKMNHQNIIKLREVV--RENNELFFI 55
M T +IVA+K+++ F WE + REI LR+++H N++KL+ +V R + L+ +
Sbjct: 49 MMTGKIVALKKVR--FDNWEPESVKFMAREILILRRLDHPNVVKLQGLVTSRMSCSLYLV 222
Query: 56 FEYMDCNLYQLIKEREKPFSEEEIR 80
F+YM+ +L L F+E ++R
Sbjct: 223 FDYMEHDLAGLAASPGIRFTEPQVR 297
>TC227247 UP|Q6T2Z8 (Q6T2Z8) Cyclin-dependent kinases CDKB, complete
Length = 1235
Score = 139 bits (351), Expect = 2e-33
Identities = 83/229 (36%), Positives = 133/229 (57%), Gaps = 12/229 (5%)
Frame = +2
Query: 23 TNLREIKALRKMNHQ-NIIKLREVVRENNE-----LFFIFEYMDCNLYQLIK---EREKP 73
T LRE+ LR ++ ++++L +V + N+ L+ +FEYMD +L + I+ + +
Sbjct: 206 TTLREVSILRMLSRDPHVVRLMDVKQGQNKEGKTVLYLVFEYMDTDLKKFIRSFRQTGQT 385
Query: 74 FSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTND--VLKIADFGLAREVS-SMP 130
+ I+ M Q+ +G++ H G HRDLKP NLL+ +LKIAD GLAR + +
Sbjct: 386 VPPQTIKSLMYQLCKGVAFCHGHGILHRDLKPHNLLMDPKTMMLKIADLGLARAFTVPIK 565
Query: 131 PYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYC 190
YT + T WYRAPEVLL + Y+ AVDMW++G I AEL T +FPG+SE+ Q+ I+
Sbjct: 566 KYTHEILTLWYRAPEVLLGATHYSMAVDMWSVGCIFAELVTKQALFPGDSELQQLLHIFR 745
Query: 191 ILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLIT 239
+LG P+ + S+L+++ + P LS +P+ +DL++
Sbjct: 746 LLGTPNE---DVWPGVSKLMNWHEYPQWNPQSLSTAVPSLDELGLDLLS 883
>TC225410 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, partial (88%)
Length = 1375
Score = 139 bits (350), Expect = 2e-33
Identities = 79/213 (37%), Positives = 119/213 (55%), Gaps = 6/213 (2%)
Frame = +3
Query: 32 RKMNHQNIIKLREVV-----RENNELFFIFEYMDCNLYQLIKEREKPFSEEEIRCFMKQM 86
R++ ++ +I LR+V+ RE N+++ E MD +L+ +I+ + SEE + F+ Q+
Sbjct: 150 RRVWNRLVIGLRDVIPPPLRREFNDVYIATELMDTDLHHIIRSNQN-LSEEHCQYFLYQI 326
Query: 87 LQGLSHMHKKGFFHRDLKPENLLVTNDV-LKIADFGLAREVSSMPPYTQYVSTRWYRAPE 145
L+GL ++H HRDLKP NLL+ ++ LKI DFGLAR T+YV TRWYRAPE
Sbjct: 327 LRGLKYIHSANVIHRDLKPSNLLLNSNCDLKIIDFGLARPTLESDFMTEYVVTRWYRAPE 506
Query: 146 VLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILGMPDSTCFTIGAN 205
+LL S YT A+D+W++G I EL P+FPG+ + QM + +LG P + N
Sbjct: 507 LLLNSSDYTSAIDVWSVGCIFMELMNKKPLFPGKDHVHQMRLLTELLGTPTEADLGLVKN 686
Query: 206 NSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLI 238
L+ + P+ AIDL+
Sbjct: 687 EDARRYIRQLPQYPRQPLAQVFPHVHPAAIDLV 785
>TC216656 weakly similar to UP|MHK_ARATH (P43294) Serine/threonine-protein
kinase MHK , partial (15%)
Length = 802
Score = 134 bits (337), Expect = 7e-32
Identities = 87/152 (57%), Positives = 95/152 (62%), Gaps = 1/152 (0%)
Frame = +1
Query: 231 SMEAIDLITARCGSVTATSFFPSCSILHKVSNFLTYTEQVNTRVPRSLSDPLELKLSNKR 290
S+EAIDLIT + S+ H QV+ VP LSDPLELKLS+KR
Sbjct: 1 SLEAIDLITQLLHWDPSRRPDADQSLQHPFF-------QVDAWVPCPLSDPLELKLSSKR 159
Query: 291 VKPNLELKLHDFGPDPDDCFLGLTLAVKPSVSNLDVVQNARQGMGEN-LV*LDAL*KCRI 349
KPNLELKL DFGPDPDDCFLGLTL VKPSVSNLDVVQN QG+ EN L D
Sbjct: 160 AKPNLELKLQDFGPDPDDCFLGLTLGVKPSVSNLDVVQNVSQGVRENVLFCSDFNDHSDQ 339
Query: 350 CYFARILMTIQTNQPDQNGVHSSAETSLSLSF 381
F +L PDQNGVH+SAETSLSLSF
Sbjct: 340 SVFWTLL------SPDQNGVHNSAETSLSLSF 417
>TC219518 similar to UP|Q9FYT8 (Q9FYT8) Cyclin-dependent kinase B1-2,
complete
Length = 1333
Score = 125 bits (315), Expect = 2e-29
Identities = 71/196 (36%), Positives = 109/196 (55%), Gaps = 8/196 (4%)
Frame = +2
Query: 52 LFFIFEYMDCNLYQLIKEREK-----PFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPE 106
L+ +FEY+D +L + I K P I+ F+ Q+ +G++H H G HRDLKP+
Sbjct: 278 LYLVFEYLDTDLKKFIDSHRKGPNPRPLPPPLIQSFLFQLCKGVAHCHSHGVLHRDLKPQ 457
Query: 107 NLLVTND--VLKIADFGLAREVS-SMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIG 163
NLL+ +LKIAD GL R + + YT + T WYRAPEVLL S Y+ VD+W++G
Sbjct: 458 NLLLDQHKGILKIADLGLGRAFTVPLKSYTHEIVTLWYRAPEVLLGSTHYSTGVDIWSVG 637
Query: 164 AILAELFTLTPIFPGESEIDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKL 223
I AE+ +FPG+SE Q+ I+ +LG P + + L D+ + P L
Sbjct: 638 CIFAEMVRRQALFPGDSEFQQLIHIFKMLGTPTEENW---PGVTSLRDWHVYPRWEPQSL 808
Query: 224 SDIIPNASMEAIDLIT 239
+ +P+ + +DL++
Sbjct: 809 AKNVPSLGPDGVDLLS 856
>TC217618 similar to GB|AAO64868.1|29028978|BT005933 At5g50860 {Arabidopsis
thaliana;} , partial (60%)
Length = 1325
Score = 121 bits (303), Expect(2) = 3e-28
Identities = 58/121 (47%), Positives = 79/121 (64%), Gaps = 3/121 (2%)
Frame = +1
Query: 78 EIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTND-VLKIADFGLAR--EVSSMPPYTQ 134
+++C+MKQ+L GL H H +G HRD+K NLL+ N+ +LKIADFGLA + P T
Sbjct: 133 QVKCYMKQLLSGLEHCHSRGVLHRDIKGSNLLIDNEGILKIADFGLATFFDPKKKHPMTS 312
Query: 135 YVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILGM 194
V T WYR PE+LL S Y VD+W+ G ILAEL P PG +E++Q++KI+ + G
Sbjct: 313 RVVTLWYRPPELLLGSTSYGVGVDLWSAGCILAELLAGKPTMPGRTEVEQLHKIFKLCGS 492
Query: 195 P 195
P
Sbjct: 493 P 495
Score = 21.9 bits (45), Expect(2) = 3e-28
Identities = 11/32 (34%), Positives = 18/32 (55%)
Frame = +2
Query: 47 RENNELFFIFEYMDCNLYQLIKEREKPFSEEE 78
R ++ L+ +FEYM+ +L L FSE +
Sbjct: 23 RISSSLYLVFEYMEHDLAGLSAAVGVKFSEPQ 118
>TC225360 homologue to UP|MSK1_MEDSA (P51137) Glycogen synthase kinase-3
homolog MsK-1 , partial (97%)
Length = 1594
Score = 122 bits (305), Expect = 3e-28
Identities = 88/289 (30%), Positives = 151/289 (51%), Gaps = 14/289 (4%)
Frame = +1
Query: 1 MRTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREV---VRENNELFF--I 55
+ T E VA+K++ + + Y N RE++ +R ++H N++ L+ E +EL+ +
Sbjct: 436 LETGETVAIKKVLQD----KRYKN-RELQTMRLLDHPNVVCLKHCFFSTTEKDELYLNLV 600
Query: 56 FEYMDCNLYQLIKEREKPFSEEE---IRCFMKQMLQGLSHMHKK-GFFHRDLKPENLLVT 111
EY+ + ++IK K ++ + Q+ + LS++H+ G HRD+KP+NLLV
Sbjct: 601 LEYVPETVNRVIKHYNKLNQRMPLIYVKLYTYQIFRALSYIHRCIGVCHRDIKPQNLLVN 780
Query: 112 NDV--LKIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAEL 169
+KI DFG A+ + P Y+ +R+YRAPE++ + YT A+D+W++G +LAEL
Sbjct: 781 PHTHQVKICDFGSAKVLVKGEPNISYICSRYYRAPELIFGATEYTTAIDVWSVGCVLAEL 960
Query: 170 FTLTPIFPGESEIDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPN 229
P+FPGES +DQ+ +I +LG P T I N +F ++ A
Sbjct: 961 MLGQPLFPGESGVDQLVEIIKVLGTP--TREEIKCMNPNYTEFKFPQIKAHPWHKIFHKR 1134
Query: 230 ASMEAIDLITARCGSVTATSFFPS--CSILHKVSN-FLTYTEQVNTRVP 275
EA+DL++ + P+ C+ L +++ F NTR+P
Sbjct: 1135MPPEAVDLVS------RLLQYSPNLRCTALDTLTHPFFDELRDPNTRLP 1263
>AI437812
Length = 421
Score = 122 bits (305), Expect = 3e-28
Identities = 58/119 (48%), Positives = 82/119 (68%), Gaps = 3/119 (2%)
Frame = +1
Query: 74 FSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTND-VLKIADFGLAR--EVSSMP 130
F+E +++C+M+Q+L+GL H H G HRD+K NLL+ N +LKIADFGLA + +
Sbjct: 19 FTEAQVKCYMQQLLRGLDHCHSCGVLHRDIKGSNLLIDNSGILKIADFGLASFFDPNQAQ 198
Query: 131 PYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIY 189
P T V T WYR PE+LL + Y AVD+W+ G ILAEL+ PI PG +E++Q++KI+
Sbjct: 199 PLTSRVVTLWYRPPELLLGATYYGTAVDLWSTGCILAELYAGKPIMPGRTEVEQLHKIF 375
>TC225359 homologue to UP|MSK1_MEDSA (P51137) Glycogen synthase kinase-3
homolog MsK-1 , complete
Length = 1668
Score = 121 bits (303), Expect = 6e-28
Identities = 79/250 (31%), Positives = 136/250 (53%), Gaps = 11/250 (4%)
Frame = +2
Query: 1 MRTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREV---VRENNELFF--I 55
+ T E VA+K++ + + Y N RE++ +R ++H N++ L+ E +EL+ +
Sbjct: 401 LETGETVAIKKVLQD----KRYKN-RELQTMRLLDHPNVVALKHCFFSTTEKDELYLNLV 565
Query: 56 FEYMDCNLYQLIKEREKPFSEEE---IRCFMKQMLQGLSHMHKK-GFFHRDLKPENLLVT 111
EY+ + ++IK K ++ + Q+ + LS++H+ G HRD+KP+NLLV
Sbjct: 566 LEYVPETVNRVIKHYNKLNQRMPLIYVKLYTYQIFRALSYIHRCIGVCHRDIKPQNLLVN 745
Query: 112 NDV--LKIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAEL 169
+K+ DFG A+ + P Y+ +R+YRAPE++ + YT A+D+W++G +LAEL
Sbjct: 746 PHTHQVKLCDFGSAKVLVKGEPNISYICSRYYRAPELIFGATEYTTAIDVWSVGCVLAEL 925
Query: 170 FTLTPIFPGESEIDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPN 229
P+FPGES +DQ+ +I +LG P T I N +F ++ A
Sbjct: 926 LLGQPLFPGESGVDQLVEIIKVLGTP--TREEIKCMNPNYTEFKFPQIKAHPWHKIFHKR 1099
Query: 230 ASMEAIDLIT 239
EA+DL++
Sbjct: 1100MPPEAVDLVS 1129
>TC206197 homologue to UP|MAPK_PEA (Q06060) Mitogen-activated protein kinase
homolog D5 , partial (59%)
Length = 940
Score = 118 bits (296), Expect = 4e-27
Identities = 65/164 (39%), Positives = 91/164 (54%), Gaps = 2/164 (1%)
Frame = +1
Query: 77 EEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVT-NDVLKIADFGLAREVSSMPPYTQY 135
E + F+ Q+L+GL ++H HRDLKP NLL+ N LKI DFGLAR S T+Y
Sbjct: 1 EHCQYFLYQILRGLKYIHSANVLHRDLKPSNLLLNANCDLKICDFGLARVTSETDFMTEY 180
Query: 136 VSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILGMP 195
V TRWYRAPE+LL S YT A+D+W++G I EL P+FPG + Q+ + ++G P
Sbjct: 181 VVTRWYRAPELLLNSSDYTAAIDVWSVGCIFMELMDRKPLFPGRDHVHQLRLLMELIGTP 360
Query: 196 DSTCFTIGANNSRLLDFVGH-EVVAPVKLSDIIPNASMEAIDLI 238
+G N ++ + + P+ EAIDL+
Sbjct: 361 SEA--DLGFLNENAKRYIRQLPLYRRQSFQEKFPHVHPEAIDLV 486
>TC205225 homologue to UP|KSG1_ARATH (P43288) Shaggy-related protein kinase
alpha (ASK-alpha) , partial (74%)
Length = 904
Score = 118 bits (296), Expect = 4e-27
Identities = 69/206 (33%), Positives = 119/206 (57%), Gaps = 11/206 (5%)
Frame = +1
Query: 1 MRTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREV---VRENNELFF--I 55
+ T E VA+K++ + + Y N RE++ +R ++H N++ L+ E +EL+ +
Sbjct: 208 LETGETVAIKKVLQD----KRYKN-RELQTMRLLDHPNVVTLKHCFFSTTEKDELYLNLV 372
Query: 56 FEYMDCNLYQLIKEREKPFSEEE---IRCFMKQMLQGLSHMHKK-GFFHRDLKPENLLVT 111
EY+ ++++I+ K ++ + Q+ + L+++H G HRD+KP+NLLV
Sbjct: 373 LEYVPETVHRVIRHYNKMNQRMPLIYVKLYFYQICRALAYIHNCIGVSHRDIKPQNLLVN 552
Query: 112 NDV--LKIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAEL 169
LKI DFG A+ + P Y+ +R+YRAPE++ + YT A+D+W+ G +L EL
Sbjct: 553 PHTHQLKICDFGSAKVLVKGEPNISYICSRYYRAPELIFGATEYTTAIDIWSAGCVLGEL 732
Query: 170 FTLTPIFPGESEIDQMYKIYCILGMP 195
P+FPGES +DQ+ +I +LG P
Sbjct: 733 LLGQPLFPGESGVDQLVEIIKVLGTP 810
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.140 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,684,102
Number of Sequences: 63676
Number of extensions: 239051
Number of successful extensions: 2383
Number of sequences better than 10.0: 558
Number of HSP's better than 10.0 without gapping: 2087
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2122
length of query: 381
length of database: 12,639,632
effective HSP length: 99
effective length of query: 282
effective length of database: 6,335,708
effective search space: 1786669656
effective search space used: 1786669656
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC136841.7


