

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.6 + phase: 0
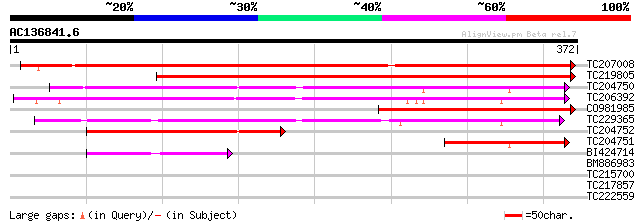
(372 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207008 558 e-159
TC219805 379 e-106
TC204750 188 4e-48
TC206392 179 2e-45
CO981985 168 3e-42
TC229365 165 3e-41
TC204752 87 2e-17
TC204751 77 1e-14
BI424714 48 6e-06
BM886983 38 0.008
TC215700 similar to GB|CAA89201.2|6469340|ATANTMR adenine nucleo... 30 1.8
TC217857 similar to UP|Q8VWW8 (Q8VWW8) Transcription factor AHAP... 30 2.3
TC222559 28 8.7
>TC207008
Length = 1489
Score = 558 bits (1439), Expect = e-159
Identities = 288/376 (76%), Positives = 319/376 (84%), Gaps = 12/376 (3%)
Frame = +2
Query: 8 KPHIGFFTRP------------ISFSSSLSFRSRVPLIKASAGASSHCESSSLNTPLLPR 55
KPH FFTRP S SS L + R P I+ASAGAS CE SSLN+PL PR
Sbjct: 209 KPHFRFFTRPDLGGGGGGGRVASSSSSCLLYHRRRPFIRASAGASP-CEFSSLNSPLEPR 385
Query: 56 TQVGKFLSGVLQNHRNLFHVAVQEELKLLADDRDAANSRMLLASESDEALLHRRIAEMKE 115
+ VGKFLS VLQNH +FHVAV EELKLLA+DRDAA++RM+L S SDEALLHRRIA++KE
Sbjct: 386 SMVGKFLSAVLQNHPQMFHVAVGEELKLLAEDRDAAHARMVLGSASDEALLHRRIAQVKE 565
Query: 116 NQCEVAVEDIMSLLIFHKFSEIRAPLVPKLSRCLYNGRLEILPSKDWELESIHTLEVLDM 175
NQC++AVED+M LLIF+KFSEIR P+VPKLS CLYNGRLEILPSKDWELESIH+ EVLDM
Sbjct: 566 NQCQIAVEDVMYLLIFYKFSEIRVPMVPKLSSCLYNGRLEILPSKDWELESIHSSEVLDM 745
Query: 176 IREHVTTVTGLKAKPSVTESWATTKVRQFLLGRIYVASILYGYFLKSVSLRYHLERNLNL 235
IREH+TTVTGL+AK SVTE WATT VRQFLL R+YVASILYGYFLKSVSLRYHLERNL+L
Sbjct: 746 IREHITTVTGLRAKSSVTECWATTHVRQFLLARVYVASILYGYFLKSVSLRYHLERNLSL 925
Query: 236 ANHDVHPGHRTNLSFKDMCPYGFEDDIFGHLSNMKPIGQGLIRQEEEIEDLKCYVMRFHP 295
ANHD+H GH+T+L MC YGF+D IFGHLSNM +GQGLIR EEEIEDLKCYVM FHP
Sbjct: 926 ANHDLHLGHKTSL----MCSYGFKDAIFGHLSNMSSLGQGLIRPEEEIEDLKCYVMSFHP 1093
Query: 296 GSLQRCAKLRSKEAVNLVRSYSSALFNSEGFDSVDSDDVILTSFSSLKRLVLEAVAFGSF 355
GSLQRCA+LRSKEAVNLV SYS ALFN++ SV++DDVILTSFSSLKRLVLEAVAFGSF
Sbjct: 1094GSLQRCARLRSKEAVNLVGSYSCALFNNKESGSVENDDVILTSFSSLKRLVLEAVAFGSF 1273
Query: 356 LWETEDYIDNVYKLKD 371
LWETEDYIDNVYKLKD
Sbjct: 1274LWETEDYIDNVYKLKD 1321
>TC219805
Length = 1061
Score = 379 bits (974), Expect = e-106
Identities = 189/275 (68%), Positives = 229/275 (82%)
Frame = +1
Query: 97 LASESDEALLHRRIAEMKENQCEVAVEDIMSLLIFHKFSEIRAPLVPKLSRCLYNGRLEI 156
++ SDEALLHRRIA++KEN+ +A+ D+M LLI +KFSEIR LVPKLS CLY+GRLEI
Sbjct: 1 ISPHSDEALLHRRIAQVKENESMIAIADVMYLLILYKFSEIRVNLVPKLSSCLYDGRLEI 180
Query: 157 LPSKDWELESIHTLEVLDMIREHVTTVTGLKAKPSVTESWATTKVRQFLLGRIYVASILY 216
LPSKDW+LESIH+LEVLD+IR+HV+TVTGL++ PSV ESW TT +RQ L R+YVASILY
Sbjct: 181 LPSKDWDLESIHSLEVLDIIRKHVSTVTGLRSNPSVRESWETTPIRQVWLARVYVASILY 360
Query: 217 GYFLKSVSLRYHLERNLNLANHDVHPGHRTNLSFKDMCPYGFEDDIFGHLSNMKPIGQGL 276
GYFLKSVSLRY+LER+L+L++HD+H GH+T SF+DM G +D + G+ S+++ + GL
Sbjct: 361 GYFLKSVSLRYNLERSLSLSDHDLHHGHKTGPSFQDMYRSGPKDVMLGNKSDIRSVWHGL 540
Query: 277 IRQEEEIEDLKCYVMRFHPGSLQRCAKLRSKEAVNLVRSYSSALFNSEGFDSVDSDDVIL 336
I QEEEIEDL CYV FHPGS +RCAKLRSKEAV+LV S+S+ALF DDVI+
Sbjct: 541 IGQEEEIEDLTCYVTGFHPGSFERCAKLRSKEAVHLVESHSNALFGDGKSGLSQHDDVIV 720
Query: 337 TSFSSLKRLVLEAVAFGSFLWETEDYIDNVYKLKD 371
TSFSSL+RLVLEAVAFGSFLWETEDYIDNVYKLKD
Sbjct: 721 TSFSSLRRLVLEAVAFGSFLWETEDYIDNVYKLKD 825
>TC204750
Length = 1791
Score = 188 bits (477), Expect = 4e-48
Identities = 112/353 (31%), Positives = 199/353 (55%), Gaps = 12/353 (3%)
Frame = +1
Query: 27 RSRVPLIKASAGASSHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLFHVAVQEELKLLAD 86
R R +++ASA +S + + + PL + +G+FLS +L++H +L AV ++L+ L
Sbjct: 259 RRRGFVVRASASSSPESDDAKI-APLQFESSIGQFLSQILKDHPHLVPAAVDQQLQQLQT 435
Query: 87 DRDAANSRMLLASESDEALLHRRIAEMKENQCEVAVEDIMSLLIFHKFSEIRAPLVPKLS 146
DRDA ++ + + +L+RRIAE+K N+ A+E+I+ L+ KF + L+P ++
Sbjct: 436 DRDAHQQNEQPSASTTDLVLYRRIAEVKANERRKALEEILYALVVQKFMDANISLIPSVT 615
Query: 147 RCLYNGRLEILPSKDWELESIHTLEVLDMIREHVTTVTGLKAKPSVTESWATTKVRQFLL 206
L +G++++ P++D +LE +H+ E +MI+ H+ + G +A + + ++ +F +
Sbjct: 616 PDL-SGKVDLWPNEDGKLELLHSHEAYEMIQNHLALILGNRA----GDLTSIAEISKFRV 780
Query: 207 GRIYVASILYGYFLKSVSLRYHLERNLNLANHDVHPGHRTNLSFKDMCPYGFEDDIFGHL 266
G++Y AS++YGYFL+ V R+ LE+ + + + + + + D E+D +
Sbjct: 781 GQVYAASVMYGYFLRRVDQRFQLEKTMKVLPNATEKENSAHQTTMDNARPSIEEDTSQVM 960
Query: 267 SNMK-------PIGQGLIRQEEEIEDLKCYVMRFHPGSLQRCAKLRSKEAVNLVRSYSSA 319
S+ + + G + L+ YVM F +LQR A +RSKEAV+++ ++ A
Sbjct: 961 SHPEVSTWPGGDVRPGGFGYGVKATRLRNYVMSFDGDTLQRYATIRSKEAVSIIEKHTEA 1140
Query: 320 LFNSEGF-----DSVDSDDVILTSFSSLKRLVLEAVAFGSFLWETEDYIDNVY 367
LF +V D+ I SF LK+LVLEAV FGSFLW+ E Y+D+ Y
Sbjct: 1141LFGRPEIVVTPEGAVSKDENIKISFGGLKKLVLEAVTFGSFLWDVESYVDSRY 1299
>TC206392
Length = 1573
Score = 179 bits (453), Expect = 2e-45
Identities = 117/386 (30%), Positives = 205/386 (52%), Gaps = 21/386 (5%)
Frame = +3
Query: 3 NCLTIKPHIGFFTR---PISFSSSLSFRSRVP-----LIKASAGASSHCESSSLNTPLLP 54
+ L + ++ F T+ +SFS R RV +++A++ E +S PL
Sbjct: 165 SALAFRSNLRFSTKLSTSVSFSKQGHARHRVGSKRGFVVRAASFTPESSEPTSKIAPLKL 344
Query: 55 RTQVGKFLSGVLQNHRNLFHVAVQEELKLLADDRDAANSRMLLASESDEALLHRRIAEMK 114
+ +G+FLS +L +H +L AV+++L+ D D + ++ + +L+RRIAE+K
Sbjct: 345 ESPIGQFLSQILISHPHLVPAAVEQQLEQFQTDCDGDKQKKEPSASGTDLVLYRRIAEVK 524
Query: 115 ENQCEVAVEDIMSLLIFHKFSEIRAPLVPKLSRCLYNGRLEILPSKDWELESIHTLEVLD 174
+ A+E+I+ L+ KF + L+P L+ ++G+++ PS+D +LE +H+ E +
Sbjct: 525 AKERRTALEEILYALVVQKFMDANISLIPSLTP-NHSGQVDSWPSEDGKLEELHSPEAYE 701
Query: 175 MIREHVTTVTGLKAKPSVTESWATTKVRQFLLGRIYVASILYGYFLKSVSLRYHLERNLN 234
MI+ H+ + G + + +S + ++ + +G++Y ASI+YGYFLK V R+ LE+ +
Sbjct: 702 MIQNHLALILGNR----LGDSTSVAQISKIRVGQVYAASIMYGYFLKWVVQRFQLEKTMK 869
Query: 235 LANHDVHPGHRTNLSFKDMCPYGFE--DDIFGH--LSNMK--PIGQGLIRQEEEIEDLKC 288
+ + + D G + + H +S + I G ++ L+
Sbjct: 870 ILPNGAEENSIQHTVVDDSRISGGDGRSHVMSHPEVSTLPGGGISSGGFGYGSKVSRLRT 1049
Query: 289 YVMRFHPGSLQRCAKLRSKEAVNLVRSYSSALF-------NSEGFDSVDSDDVILTSFSS 341
YVM F +LQR A +RSKEA++++ ++ ALF EG +D+ I SF
Sbjct: 1050YVMSFDSETLQRYATIRSKEALSIIEKHTEALFGRPEIVVTPEGVIDSLTDESIKISFGG 1229
Query: 342 LKRLVLEAVAFGSFLWETEDYIDNVY 367
LKRLVLEA+ FGSFLW+ E Y+D+ Y
Sbjct: 1230LKRLVLEAITFGSFLWDVESYVDSRY 1307
>CO981985
Length = 903
Score = 168 bits (426), Expect = 3e-42
Identities = 85/129 (65%), Positives = 102/129 (78%)
Frame = -2
Query: 243 GHRTNLSFKDMCPYGFEDDIFGHLSNMKPIGQGLIRQEEEIEDLKCYVMRFHPGSLQRCA 302
GH+ + SF DM G +D +FG+ S+++ + GLI QEEEIEDLKCYV FHPGS +RCA
Sbjct: 899 GHKISPSFHDMYHSGAKDVMFGNKSDIQSVWHGLIGQEEEIEDLKCYVTGFHPGSFERCA 720
Query: 303 KLRSKEAVNLVRSYSSALFNSEGFDSVDSDDVILTSFSSLKRLVLEAVAFGSFLWETEDY 362
KLRSKEAV+LV S+S+ALF DD+I+TSFSSL+RLVLEAVAFGSFLWETEDY
Sbjct: 719 KLRSKEAVHLVESHSNALFGDGKSGLSQHDDIIVTSFSSLRRLVLEAVAFGSFLWETEDY 540
Query: 363 IDNVYKLKD 371
ID+VYKLKD
Sbjct: 539 IDSVYKLKD 513
>TC229365
Length = 1346
Score = 165 bits (418), Expect = 3e-41
Identities = 111/358 (31%), Positives = 189/358 (52%), Gaps = 10/358 (2%)
Frame = +2
Query: 17 PISFSSSLSFRSRVPLIKASAGASSHCESSSLNTPLLPRTQVGKFLSGVLQNHRNLFHVA 76
P ++ +FR+R + +ASA + + PL + VG+ L + H +L A
Sbjct: 314 PKHYNELYNFRARGLVARASADPRDNLVPFA---PLQFESPVGQLLEQISNTHPHLLPAA 484
Query: 77 VQEELKLLADDRDAANSRMLLASESDEALLHRRIAEMKENQCEVAVEDIMSLLIFHKFSE 136
+ ++L+ L RDA N +S+S L++RIAE+KE + +E+I+ I HKF E
Sbjct: 485 IDQQLENLQTARDAQNE----SSDSSLDSLYKRIAEVKEKEKRTTLEEILYCSIVHKFLE 652
Query: 137 IRAPLVPKLSRCLY-NGRLEILPSKDWELESIHTLEVLDMIREHVTTVTGLKAKPSVTES 195
++PK+S GR+++ P+++ +LE +H+ E +MI+ H++ V G + V
Sbjct: 653 NNISMIPKISATSDPTGRVDLWPNQELKLEGVHSPEAFEMIQSHLSLVLGDRL---VGPL 823
Query: 196 WATTKVRQFLLGRIYVASILYGYFLKSVSLRYHLERNLNLANHDVHPGHRTNLSFKDMCP 255
++ + LG++Y ASI+YGYFLK + R+ LER++ D SF + P
Sbjct: 824 QTVVQISKIKLGKLYAASIMYGYFLKRIDERFQLERSMGTLPKDFGKAK----SFDEPSP 991
Query: 256 --YGFEDDIFGHLSNMKPIGQGLIRQEEEIEDLKCYVMRFHPGSLQRCAKLRSKEAVNLV 313
++ D + + + E L+ YVM+ +LQR A +RSKEA++L+
Sbjct: 992 GIKLWDPDSLIIVHDYDNDSDHMDTDEGRSFRLRAYVMQLDAETLQRLATVRSKEAISLI 1171
Query: 314 RSYSSALF-------NSEGFDSVDSDDVILTSFSSLKRLVLEAVAFGSFLWETEDYID 364
+ ALF + +G +D+++ +FS L LVLEA+AFGSFLW+ E+Y++
Sbjct: 1172EKQTQALFGRPDIRVSEDGSIETSNDELLSLTFSGLTMLVLEALAFGSFLWDKENYVE 1345
>TC204752
Length = 792
Score = 86.7 bits (213), Expect = 2e-17
Identities = 45/131 (34%), Positives = 81/131 (61%)
Frame = +1
Query: 51 PLLPRTQVGKFLSGVLQNHRNLFHVAVQEELKLLADDRDAANSRMLLASESDEALLHRRI 110
PL + +G+FLS +L++H +L AV ++L L DRDA S + +L+RRI
Sbjct: 307 PLQLESPIGQFLSQILKDHPHLVPAAVDQQLHQLQTDRDAHLQNQQQPSSPTDLVLYRRI 486
Query: 111 AEMKENQCEVAVEDIMSLLIFHKFSEIRAPLVPKLSRCLYNGRLEILPSKDWELESIHTL 170
AE+K N+ A+E+I+ L+ KF + L+P ++ L +G++++ P++D +LE +H+
Sbjct: 487 AEVKANERRKALEEILYALVVQKFMDANISLIPSVTPDL-SGKVDLWPNEDGKLEQLHSD 663
Query: 171 EVLDMIREHVT 181
E +MI+ H++
Sbjct: 664 EAYEMIQNHLS 696
>TC204751
Length = 795
Score = 77.4 bits (189), Expect = 1e-14
Identities = 41/87 (47%), Positives = 55/87 (63%), Gaps = 5/87 (5%)
Frame = +1
Query: 286 LKCYVMRFHPGSLQRCAKLRSKEAVNLVRSYSSALFNSEGF-----DSVDSDDVILTSFS 340
L+ YVM F +LQR A +RSKEAV+++ ++ ALF +V D+ I SF
Sbjct: 274 LRNYVMSFDGDTLQRYAAIRSKEAVSIIEKHTEALFGRPEIVVTPEGAVSKDENIKISFG 453
Query: 341 SLKRLVLEAVAFGSFLWETEDYIDNVY 367
LK+LVLEAV FGSFLW+ E Y+D+ Y
Sbjct: 454 GLKKLVLEAVTFGSFLWDVESYVDSRY 534
>BI424714
Length = 353
Score = 48.1 bits (113), Expect = 6e-06
Identities = 28/96 (29%), Positives = 50/96 (51%)
Frame = +3
Query: 51 PLLPRTQVGKFLSGVLQNHRNLFHVAVQEELKLLADDRDAANSRMLLASESDEALLHRRI 110
PL + VG+ L + H +L + ++L+ L + RDA S+ + L++RI
Sbjct: 48 PLQLESPVGQLLEKISNTHPHLLTAVIDQQLENLQNVRDAQKE-----SDPSQDSLYKRI 212
Query: 111 AEMKENQCEVAVEDIMSLLIFHKFSEIRAPLVPKLS 146
AE+K+ + + + I+ I HKF E ++PK+S
Sbjct: 213 AEVKDKEKRIILPHII*C*IVHKFLENNISMIPKIS 320
>BM886983
Length = 421
Score = 37.7 bits (86), Expect = 0.008
Identities = 20/58 (34%), Positives = 32/58 (54%)
Frame = +2
Query: 51 PLLPRTQVGKFLSGVLQNHRNLFHVAVQEELKLLADDRDAANSRMLLASESDEALLHR 108
PL + +G+FLS +L++H +L AV ++L L DRDA S + +L+R
Sbjct: 188 PLQLESPIGQFLSQILKDHPHLVPAAVDQQLHQLQTDRDAHLQNQQQPSSPTDLVLYR 361
>TC215700 similar to GB|CAA89201.2|6469340|ATANTMR adenine nucleotide
translocase {Arabidopsis thaliana;} , partial (19%)
Length = 855
Score = 30.0 bits (66), Expect = 1.8
Identities = 19/50 (38%), Positives = 26/50 (52%), Gaps = 3/50 (6%)
Frame = -3
Query: 5 LTIKPHIGFFT---RPISFSSSLSFRSRVPLIKASAGASSHCESSSLNTP 51
L++ G T R ISFSSS S R+ V + A+S E+ S +TP
Sbjct: 343 LSVSATTGILTAAARSISFSSSSSLRNAVNWVSKDFAAASQAETISRSTP 194
>TC217857 similar to UP|Q8VWW8 (Q8VWW8) Transcription factor AHAP2, partial
(9%)
Length = 684
Score = 29.6 bits (65), Expect = 2.3
Identities = 12/35 (34%), Positives = 22/35 (62%)
Frame = -1
Query: 213 SILYGYFLKSVSLRYHLERNLNLANHDVHPGHRTN 247
+++YG FLK + +++ NLN + +H H+TN
Sbjct: 519 TVMYGLFLKKGEVGHNINNNLNCIHSYIH-AHKTN 418
>TC222559
Length = 674
Score = 27.7 bits (60), Expect = 8.7
Identities = 20/63 (31%), Positives = 27/63 (42%)
Frame = -2
Query: 220 LKSVSLRYHLERNLNLANHDVHPGHRTNLSFKDMCPYGFEDDIFGHLSNMKPIGQGLIRQ 279
L +SL LE N N + HRT + PY +F LS +PIG + R
Sbjct: 454 LTIISLEKQLEYNQRTKNR-IEYIHRTKNKITNRNPYSLLFFVFFFLSRQQPIGLRIQRG 278
Query: 280 EEE 282
E+
Sbjct: 277 REK 269
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.136 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,579,672
Number of Sequences: 63676
Number of extensions: 228978
Number of successful extensions: 1133
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 1116
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1121
length of query: 372
length of database: 12,639,632
effective HSP length: 99
effective length of query: 273
effective length of database: 6,335,708
effective search space: 1729648284
effective search space used: 1729648284
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC136841.6


