

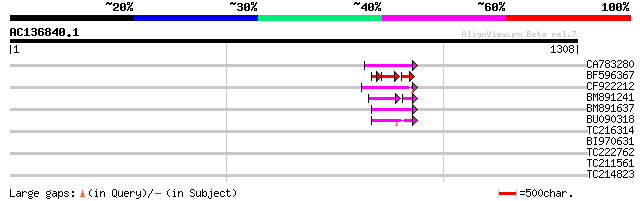
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136840.1 + phase: 0 /pseudo
(1308 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polym... 63 1e-09
BF596367 40 2e-09
CF922212 59 1e-08
BM891241 44 2e-05
BM891637 45 2e-04
BU090318 weakly similar to GP|9743345|gb| F21J9.30 {Arabidopsis ... 44 4e-04
TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {A... 41 0.003
BI970631 34 0.48
TC222762 32 1.4
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 32 1.4
TC214823 homologue to UP|RS8_PRUAR (O81361) 40S ribosomal protei... 26 2.2
>CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polymerase
homolog T24M8.8 - Arabidopsis thaliana, partial (4%)
Length = 456
Score = 62.8 bits (151), Expect = 1e-09
Identities = 33/125 (26%), Positives = 59/125 (46%), Gaps = 4/125 (3%)
Frame = +2
Query: 819 IESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQGWRILHNE 878
+ES+ RF WG + RKI W+ VC K GG+G + + N +L K W + + +
Sbjct: 41 LESLQRRFLWGGEADSRKIAWVNWKTVCLPKAKGGLGIKDLRTFNTTLLGKWRWDLFYIQ 220
Query: 879 ELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHA----KFEVIDKVGVWRVGDGRNI 934
+ + L+ +Y + S G S W+ ++ + + + +W+VG G I
Sbjct: 221 QEPWAKVLQSKYGGWRALEEGSSGSKDSAWWKDLIKTQQLQRNIPLKRETIWKVGGGDRI 400
Query: 935 RVWKD 939
+ W+D
Sbjct: 401 KFWED 415
>BF596367
Length = 419
Score = 40.4 bits (93), Expect(3) = 2e-09
Identities = 18/43 (41%), Positives = 27/43 (61%)
Frame = +1
Query: 857 RTFKELNLAMLSKQGWRILHNEELLLS*CLKGRYFPRGDFLKA 899
R K + AML+KQ WR++ N++ L++* L +YFP L A
Sbjct: 58 RRIKAFHKAMLAKQMWRLIDNKDALVT*VLTAKYFPNSSLLDA 186
Score = 31.2 bits (69), Expect(3) = 2e-09
Identities = 11/32 (34%), Positives = 23/32 (71%)
Frame = +3
Query: 903 FNPSYTWRSIMHAKFEVIDKVGVWRVGDGRNI 934
+ P+Y+W S+ +K ++ + +WR+GDGR++
Sbjct: 198 YMPNYSWSSV**SK-DLAEHGAIWRIGDGRSV 290
Score = 28.9 bits (63), Expect(3) = 2e-09
Identities = 10/24 (41%), Positives = 17/24 (70%)
Frame = +2
Query: 835 RKIHWIR*DEVCKNKKVGGMGFRT 858
+K+HW+ D++ K+KK+GG T
Sbjct: 8 QKLHWLSFDKLTKSKKIGGSKLST 79
>CF922212
Length = 445
Score = 58.9 bits (141), Expect = 1e-08
Identities = 37/137 (27%), Positives = 62/137 (45%), Gaps = 8/137 (5%)
Frame = -2
Query: 811 VPNG*CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQ 870
VPN + + + F WG G++KI W+ D VC K+ G+G R ++ N A+L K+
Sbjct: 441 VPNKVLDKLVRLQQWFLWGGGLGQKKIAWVNWDSVCLPKEKEGLGNRDLRKFNYALLGKR 262
Query: 871 GWRILHNEELLLS*CLKGRYFPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVW---- 926
W + H++ L + L +Y + + + S+ W+ I D G W
Sbjct: 261 RWNLFHHQGELGARVLDSKYKRWRNLDEERRVKSESFWWQEISFITHSTED--GSWFEKR 88
Query: 927 ----RVGDGRNIRVWKD 939
R+G +R W+D
Sbjct: 87 TKTGRLGCRAKVRFWED 37
>BM891241
Length = 407
Score = 44.3 bits (103), Expect(2) = 2e-05
Identities = 21/73 (28%), Positives = 37/73 (49%)
Frame = -2
Query: 829 GSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQGWRILHNEELLLS*CLKG 888
G +KI W++ + VC K GG+G + + N+A+L K W + +++ L + +
Sbjct: 406 GGDHDHKKIPWVKWEVVCLPKTKGGLGIKDISKFNIALLGKWIWALASDQQQLWARIINS 227
Query: 889 RYFPRGDFLKASV 901
+Y DF SV
Sbjct: 226 KYGGWSDFQYNSV 188
Score = 23.9 bits (50), Expect(2) = 2e-05
Identities = 11/38 (28%), Positives = 16/38 (41%), Gaps = 4/38 (10%)
Frame = -3
Query: 906 SYTWRSIM----HAKFEVIDKVGVWRVGDGRNIRVWKD 939
SY WR + + + + W+VG G I W D
Sbjct: 174 SYWWRDLRKFYHQSDHSIFHQYMSWKVGCGDKINFWTD 61
>BM891637
Length = 427
Score = 45.4 bits (106), Expect = 2e-04
Identities = 30/110 (27%), Positives = 45/110 (40%), Gaps = 4/110 (3%)
Frame = -1
Query: 835 RKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQGWRILHNEELLLS*CLKGRYFPRG 894
+KI WI + C + VGG+G + K LN A+L K W + H L + L +Y
Sbjct: 418 KKIAWISWRQCCASGDVGGLGIKDIKILNNALLIKWKWLMFHQPHQLWNRILISKYKGWR 239
Query: 895 DFLKASVGFNPSYTWRSI----MHAKFEVIDKVGVWRVGDGRNIRVWKDN 940
+ + S W + H W+VG G I W+D+
Sbjct: 238 GLDQGPQKYYFSPWWADLRAINQHQSMIAASNQFCWKVGRGDQILFWEDS 89
>BU090318 weakly similar to GP|9743345|gb| F21J9.30 {Arabidopsis thaliana},
partial (1%)
Length = 422
Score = 44.3 bits (103), Expect = 4e-04
Identities = 28/110 (25%), Positives = 47/110 (42%), Gaps = 6/110 (5%)
Frame = +3
Query: 836 KIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQGWRILHNEELLLS*CLKGRY----- 890
K+HW +C KK GG+G R +N ++L K + L ++ +Y
Sbjct: 3 KVHWTSWKALCSPKKSGGLGLRLLSHMNKSLLMKST*NMCTKPNDL*VRVVRSKYK*GKD 182
Query: 891 -FPRGDFLKASVGFNPSYTWRSIMHAKFEVIDKVGVWRVGDGRNIRVWKD 939
P D K F W+ I+H ++ ++G+GR++ WKD
Sbjct: 183 ILP*IDPKKTGTNF-----WQGIVHCWEKITHDNIARKIGNGRSVHFWKD 317
>TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {Arabidopsis
thaliana;} , partial (18%)
Length = 759
Score = 41.2 bits (95), Expect = 0.003
Identities = 17/71 (23%), Positives = 33/71 (45%)
Frame = +2
Query: 811 VPNG*CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGMGFRTFKELNLAMLSKQ 870
+P + + ++ +F WG Q +I W++ ++C K GG+G + N A+ +
Sbjct: 74 IPQRIVDKLVTLQRQFLWGGTQHHNRIPWVKWADICNPKIDGGLGIKDLSNFNAALRGRW 253
Query: 871 GWRILHNEELL 881
W + N L
Sbjct: 254 IWGLASNHNQL 286
>BI970631
Length = 775
Score = 33.9 bits (76), Expect = 0.48
Identities = 15/39 (38%), Positives = 20/39 (50%), Gaps = 4/39 (10%)
Frame = +1
Query: 906 SYTWRSI----MHAKFEVIDKVGVWRVGDGRNIRVWKDN 940
S+ WR + H F++I W+ G G IR WKDN
Sbjct: 493 SHWWRDLRKIYQHNDFKIIHXXMAWKAGCGDKIRFWKDN 609
>TC222762
Length = 879
Score = 32.3 bits (72), Expect = 1.4
Identities = 11/28 (39%), Positives = 20/28 (71%)
Frame = +3
Query: 889 RYFPRGDFLKASVGFNPSYTWRSIMHAK 916
+++ GDFL +++G+ PS WRSI ++
Sbjct: 312 KHYSLGDFLDSNLGYGPSLVWRSIWSSR 395
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 32.3 bits (72), Expect = 1.4
Identities = 14/44 (31%), Positives = 24/44 (53%)
Frame = +1
Query: 811 VPNG*CEHIESMISRFWWGSKQGERKIHWIR*DEVCKNKKVGGM 854
VP + + + F WG ++KI WIR +++C K+ GG+
Sbjct: 778 VPQSVVDKLVKI*RTFLWGGGPDQKKISWIRWEKLCLPKERGGI 909
>TC214823 homologue to UP|RS8_PRUAR (O81361) 40S ribosomal protein S8, partial
(93%)
Length = 940
Score = 25.8 bits (55), Expect(2) = 2.2
Identities = 9/12 (75%), Positives = 9/12 (75%)
Frame = +3
Query: 1236 NC*GL*GCCCCC 1247
NC G GCCCCC
Sbjct: 489 NCRG--GCCCCC 518
Score = 24.3 bits (51), Expect(2) = 2.2
Identities = 8/19 (42%), Positives = 10/19 (52%)
Frame = +3
Query: 1243 CCCCCSMLENNDSSKFRYG 1261
CCCCC + SS + G
Sbjct: 507 CCCCCGRSQKE*SSAEKIG 563
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.358 0.160 0.572
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 61,045,896
Number of Sequences: 63676
Number of extensions: 915857
Number of successful extensions: 13540
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 6715
Number of HSP's successfully gapped in prelim test: 313
Number of HSP's that attempted gapping in prelim test: 4451
Number of HSP's gapped (non-prelim): 8949
length of query: 1308
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1200
effective length of database: 5,762,624
effective search space: 6915148800
effective search space used: 6915148800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC136840.1


