

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136507.3 + phase: 0 /pseudo
(491 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
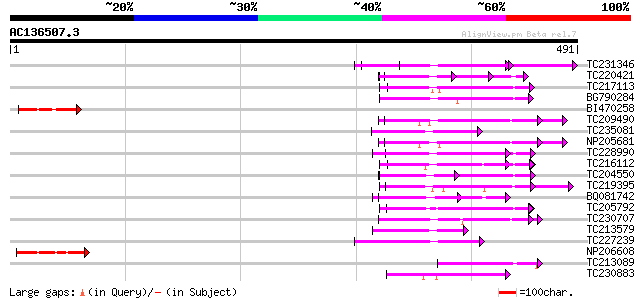
Score E
Sequences producing significant alignments: (bits) Value
TC231346 weakly similar to UP|Q6QM04 (Q6QM04) LRR protein WM1.2,... 105 5e-23
TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell... 68 8e-12
TC217113 similar to UP|Q708X5 (Q708X5) Leucine rich repeat prote... 67 2e-11
BG790284 weakly similar to GP|3894383|gb|A disease resistance pr... 66 4e-11
BI470258 65 5e-11
TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment),... 65 9e-11
TC235081 weakly similar to UP|O65580 (O65580) Receptor protein k... 62 4e-10
NP205681 receptor protein kinase-like protein 62 8e-10
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 59 6e-09
TC216112 similar to UP|Q8GTD6 (Q8GTD6) Polygalacturonase inhibit... 58 1e-08
TC204550 UP|Q8L3Y5 (Q8L3Y5) Receptor-like kinase RHG1, complete 57 1e-08
TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, part... 57 2e-08
BQ081742 57 2e-08
TC205792 similar to UP|Q93X72 (Q93X72) Leucine-rich repeat resis... 56 3e-08
TC230707 similar to UP|Q12937 (Q12937) BS-84, partial (4%) 56 4e-08
TC213579 weakly similar to UP|O50024 (O50024) Hcr9-4E, partial (... 55 9e-08
TC227239 similar to UP|Q6NQP4 (Q6NQP4) At3g43740, partial (79%) 54 2e-07
NP206608 unknown 53 3e-07
TC213089 similar to PIR|T05606|T05606 protein kinase homolog F9D... 53 4e-07
TC230883 UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, complete 53 4e-07
>TC231346 weakly similar to UP|Q6QM04 (Q6QM04) LRR protein WM1.2, partial
(14%)
Length = 1790
Score = 105 bits (262), Expect = 5e-23
Identities = 70/195 (35%), Positives = 104/195 (52%), Gaps = 2/195 (1%)
Frame = +1
Query: 299 IYRVTIWQA-VSMVS*SVQS*HSLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLP 357
IY VT ++ + + S+Q+ +L +++N L G +P S G L L LDLS+N + +P
Sbjct: 1 IYTVTFYRGKIPQIISSLQNIKNLDLQNNQLSGPLPDSLGQLKHLEVLDLSNNTFTCPIP 180
Query: 358 VMLHNLSVGCAKNSLKELYLASNQIIGTVPDMSGF-SSLENMFLYENLLNGTILKNSTFP 416
NLS SL+ L LA N++ GT+P F +L+ + L N L G +
Sbjct: 181 SPFANLS------SLRTLNLAHNRLNGTIPKSFEFLKNLQVLNLGANSLTGDVPVTLGTL 342
Query: 417 YRLANLYLDSNDLDGVITDSHFGNMSMLKYLSLSSNSLALKFSENWVPPFQLSTIYLRSC 476
L L L SN L+G I +S+F + LK L LS +L L + W PPFQL + L S
Sbjct: 343 SNLVTLDLSSNLLEGSIKESNFVKLFTLKELRLSWTNLFLSVNSGWAPPFQLEYVLLSSF 522
Query: 477 TLGPSFPKWIRSQKN 491
+GP FP+W++ Q +
Sbjct: 523 GIGPKFPEWLKRQSS 567
Score = 47.0 bits (110), Expect = 2e-05
Identities = 41/130 (31%), Positives = 59/130 (44%), Gaps = 1/130 (0%)
Frame = +1
Query: 305 WQAVSMVS*SVQS*HSLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLS 364
WQA+ V+ + SN++ G IP S G L L SL L N+ S +P L N S
Sbjct: 916 WQALVHVN----------LGSNNMSGEIPNSMGYLSQLESLLLDDNRFSGYIPSTLQNCS 1065
Query: 365 VGCAKNSLKELYLASNQIIGTVPD-MSGFSSLENMFLYENLLNGTILKNSTFPYRLANLY 423
++K + + +NQ+ T+PD M L + L N NG+I + L L
Sbjct: 1066 ------TMKFIDMGNNQLSDTIPDWMWEMQYLMVLRLRSNNFNGSITQKMCQLSSLIVLD 1227
Query: 424 LDSNDLDGVI 433
+N L G I
Sbjct: 1228 HGNNSLSGSI 1257
Score = 41.6 bits (96), Expect = 8e-04
Identities = 34/100 (34%), Positives = 54/100 (54%), Gaps = 1/100 (1%)
Frame = +1
Query: 338 SLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGTVP-DMSGFSSLE 396
+L +R +DLSSNKLS +P + LS +L+ L L+ N + G +P DM LE
Sbjct: 1411 NLILVRMIDLSSNKLSGAIPSEISKLS------ALRFLNLSRNHLSGEIPNDMGKMKLLE 1572
Query: 397 NMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDS 436
++ L N ++G I ++ + L+ L L ++L G I S
Sbjct: 1573 SLDLSLNNISGQIPQSLSDLSFLSFLNLSYHNLSGRIPTS 1692
>TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell wall
protein gp1 precursor (Hydroxyproline-rich glycoprotein
1), partial (13%)
Length = 1409
Score = 68.2 bits (165), Expect = 8e-12
Identities = 50/126 (39%), Positives = 72/126 (56%), Gaps = 1/126 (0%)
Frame = +2
Query: 325 SNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIG 384
+N L+G IP S L SL+ L+LSSN L+ ++P + +L SLK L L+SN G
Sbjct: 668 NNRLKGNIPPSITMLDSLQFLNLSSNSLAGEIPASIGDLI------SLKNLSLSSNSFSG 829
Query: 385 TVPD-MSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMSM 443
++PD +S SL +M L N LNGTI K + L +L L +N+L GV+ N+S
Sbjct: 830 SIPDSVSALPSLIHMDLSSNQLNGTIPKFISQMKSLKHLNLANNNLHGVVP----FNLSF 997
Query: 444 LKYLSL 449
+K L +
Sbjct: 998 IKRLEV 1015
Score = 59.3 bits (142), Expect = 4e-09
Identities = 41/100 (41%), Positives = 57/100 (57%), Gaps = 1/100 (1%)
Frame = +2
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
L + SNSL G IP S G L SL++L LSSN S +P + L SL + L+SN
Sbjct: 728 LNLSSNSLAGEIPASIGDLISLKNLSLSSNSFSGSIPDSVSALP------SLIHMDLSSN 889
Query: 381 QIIGTVPD-MSGFSSLENMFLYENLLNGTILKNSTFPYRL 419
Q+ GT+P +S SL+++ L N L+G + N +F RL
Sbjct: 890 QLNGTIPKFISQMKSLKHLNLANNNLHGVVPFNLSFIKRL 1009
Score = 47.8 bits (112), Expect = 1e-05
Identities = 28/68 (41%), Positives = 39/68 (57%)
Frame = +2
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
+L + SNS G IP S +L SL +DLSSN+L+ +P + + SLK L LA+
Sbjct: 797 NLSLSSNSFSGSIPDSVSALPSLIHMDLSSNQLNGTIPKFISQM------KSLKHLNLAN 958
Query: 380 NQIIGTVP 387
N + G VP
Sbjct: 959 NNLHGVVP 982
>TC217113 similar to UP|Q708X5 (Q708X5) Leucine rich repeat protein
precursor, partial (96%)
Length = 1553
Score = 66.6 bits (161), Expect = 2e-11
Identities = 54/146 (36%), Positives = 71/146 (47%), Gaps = 19/146 (13%)
Frame = +3
Query: 328 LEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLS----VGCAKN------------- 370
+ G IP +L SLR LDL NKLS ++P + LS + A N
Sbjct: 357 IAGEIPTCVTALPSLRILDLIGNKLSGEIPADVGKLSRLTVLNLADNALSGKIPASITQL 536
Query: 371 -SLKELYLASNQIIGTVP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSND 428
SLK L L++NQ+ G +P D L M L N L G I + + YRLA+L L +N
Sbjct: 537 GSLKHLDLSNNQLCGEIPEDFGNLGMLSRMLLSRNQLTGKIPVSVSKIYRLADLDLSANR 716
Query: 429 LDGVITDSHFGNMSMLKYLSLSSNSL 454
L G + G M +L L+L SNSL
Sbjct: 717 LSGSV-PFELGTMPVLSTLNLDSNSL 791
Score = 51.6 bits (122), Expect = 8e-07
Identities = 43/135 (31%), Positives = 67/135 (48%), Gaps = 1/135 (0%)
Frame = +3
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
L + +N L G IP+ FG+L L + LS N+L+ +PV + + L +L L++N
Sbjct: 552 LDLSNNQLCGEIPEDFGNLGMLSRMLLSRNQLTGKIPVSVSKI------YRLADLDLSAN 713
Query: 381 QIIGTVP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFG 439
++ G+VP ++ L + L N L G I + + L L N +G I D FG
Sbjct: 714 RLSGSVPFELGTMPVLSTLNLDSNSLEGLIPSSLLSNGGMGILNLSRNGFEGSIPDV-FG 890
Query: 440 NMSMLKYLSLSSNSL 454
+ S L LS N+L
Sbjct: 891 SHSYFMALDLSFNNL 935
Score = 38.5 bits (88), Expect = 0.007
Identities = 30/92 (32%), Positives = 42/92 (45%), Gaps = 1/92 (1%)
Frame = +3
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
L + N EG IP FGS +LDLS N L +P L + + L L+ N
Sbjct: 840 LNLSRNGFEGSIPDVFGSHSYFMALDLSFNNLKGRVPSSL------ASAKFIGHLDLSHN 1001
Query: 381 QIIGTVPDMSGFSSLE-NMFLYENLLNGTILK 411
+ G++P + F LE + F + L G LK
Sbjct: 1002HLCGSIPLGAPFDHLEASSFTSNDCLCGNPLK 1097
Score = 35.0 bits (79), Expect = 0.076
Identities = 24/65 (36%), Positives = 30/65 (45%)
Frame = +3
Query: 4 VSGSCIEKERHALLELKSGLVLDDTYLLPSWDTKSDDCCAWEGIGCRNQTGHVEILDLNS 63
VSG C +R ALL + L L SW T S+ C W GI C TG V ++L
Sbjct: 63 VSG-CSPSDRAALLAFRKALSEPYLGLFNSW-TGSNCCLDWYGISCDATTGRVTDINLRG 236
Query: 64 DQFGP 68
+ P
Sbjct: 237 ESEDP 251
Score = 33.5 bits (75), Expect = 0.22
Identities = 33/113 (29%), Positives = 54/113 (47%), Gaps = 1/113 (0%)
Frame = +3
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
+L + SNSLEG IP S S + L+LS N +P + + S A L L+
Sbjct: 765 TLNLDSNSLEGLIPSSLLSNGGMGILNLSRNGFEGSIPDVFGSHSYFMA------LDLSF 926
Query: 380 NQIIGTVP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDG 431
N + G VP ++ + ++ L N L G+I + F + A+ + ++ L G
Sbjct: 927 NNLKGRVPSSLASAKFIGHLDLSHNHLCGSIPLGAPFDHLEASSFTSNDCLCG 1085
>BG790284 weakly similar to GP|3894383|gb|A disease resistance protein
{Lycopersicon esculentum}, partial (6%)
Length = 421
Score = 65.9 bits (159), Expect = 4e-11
Identities = 47/136 (34%), Positives = 72/136 (52%), Gaps = 3/136 (2%)
Frame = +2
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
L + N++ G +P S G + +L LDLSSN LS +P L NL+ ++ LY+ +N
Sbjct: 35 LDVHDNAIMGQVPNSIGQMQALEKLDLSSNMLSGSIPSSLTNLT------AISVLYMDTN 196
Query: 381 QIIGTV--PDMSG-FSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSH 437
+ GT+ P SG SL + L+ N L+G I + + L + L +N ++G + S
Sbjct: 197 YLEGTIPFPSRSGEMPSLGFLRLHNNHLSGNIPPSFGYLVSLKRVSLSNNKIEGALPSS- 373
Query: 438 FGNMSMLKYLSLSSNS 453
GN+ L L LS NS
Sbjct: 374 LGNLHSLTELYLSDNS 421
>BI470258
Length = 415
Score = 65.5 bits (158), Expect = 5e-11
Identities = 33/55 (60%), Positives = 39/55 (70%)
Frame = +1
Query: 8 CIEKERHALLELKSGLVLDDTYLLPSWDTKSDDCCAWEGIGCRNQTGHVEILDLN 62
CIE+ER ALL+ K+ LV DD +L SW T DCC WEGI C N TGHV +LDL+
Sbjct: 133 CIEREREALLQFKAALV-DDYGMLSSWTTA--DCCQWEGIRCTNLTGHVLMLDLH 288
>TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment), complete
Length = 3298
Score = 64.7 bits (156), Expect = 9e-11
Identities = 49/138 (35%), Positives = 72/138 (51%), Gaps = 1/138 (0%)
Frame = +1
Query: 325 SNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIG 384
+N+ EGGIP FGS+ SLR LDLSS LS ++P L NL+ +L L+L N + G
Sbjct: 772 NNAYEGGIPPEFGSMKSLRYLDLSSCNLSGEIPPSLANLT------NLDTLFLQINNLTG 933
Query: 385 TVP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMSM 443
T+P ++S SL ++ L N L G I + + L + N+L G + S G +
Sbjct: 934 TIPSELSAMVSLMSLDLSINDLTGEIPMSFSQLRNLTLMNFFQNNLRGSV-PSFVGELPN 1110
Query: 444 LKYLSLSSNSLALKFSEN 461
L+ L L N+ + N
Sbjct: 1111LETLQLWDNNFSFVLPPN 1164
Score = 57.0 bits (136), Expect = 2e-08
Identities = 57/184 (30%), Positives = 84/184 (44%), Gaps = 20/184 (10%)
Frame = +1
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLS--------------EDLPVMLHN--- 362
+L + N+L G +PK +L SL+ L++S N S E L V +N
Sbjct: 391 NLTVSQNNLTGVLPKELAALTSLKHLNISHNVFSGHFPGQIILPMTKLEVLDVYDNNFTG 570
Query: 363 -LSVGCAK-NSLKELYLASNQIIGTVPDM-SGFSSLENMFLYENLLNGTILKNSTFPYRL 419
L V K LK L L N G++P+ S F SLE + L N L+G I K+ + L
Sbjct: 571 PLPVELVKLEKLKYLKLDGNYFSGSIPESYSEFKSLEFLSLSTNSLSGKIPKSLSKLKTL 750
Query: 420 ANLYLDSNDLDGVITDSHFGNMSMLKYLSLSSNSLALKFSENWVPPFQLSTIYLRSCTLG 479
L L N+ FG+M L+YL LSS +L+ + + L T++L+ L
Sbjct: 751 RYLKLGYNNAYEGGIPPEFGSMKSLRYLDLSSCNLSGEIPPSLANLTNLDTLFLQINNLT 930
Query: 480 PSFP 483
+ P
Sbjct: 931 GTIP 942
Score = 34.7 bits (78), Expect = 0.100
Identities = 23/71 (32%), Positives = 35/71 (48%)
Frame = +1
Query: 323 IKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQI 382
+ N LEG IPK +L L ++S N++S +P + + SL L L++N
Sbjct: 1699 LSRNMLEGKIPKGIKNLTDLSIFNVSINQISGPVPEEIRFML------SLTTLDLSNNNF 1860
Query: 383 IGTVPDMSGFS 393
IG VP F+
Sbjct: 1861 IGKVPTGGQFA 1893
>TC235081 weakly similar to UP|O65580 (O65580) Receptor protein kinase-like
protein, partial (13%)
Length = 1002
Score = 62.4 bits (150), Expect = 4e-10
Identities = 39/97 (40%), Positives = 53/97 (54%), Gaps = 1/97 (1%)
Frame = +1
Query: 314 SVQS*HSLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLK 373
S+ S SLI +L G IPK G L L LDLS N LS ++P L C L+
Sbjct: 616 SLLSLTSLIFTGTNLTGSIPKEIGELVELGYLDLSDNALSGEIPSEL------CYLPKLE 777
Query: 374 ELYLASNQIIGTVPDMSG-FSSLENMFLYENLLNGTI 409
EL+L SN ++G++P G + L+ + LY+N L G I
Sbjct: 778 ELHLNSNDLVGSIPVAIGNLTKLQKLILYDNQLGGKI 888
Score = 28.9 bits (63), Expect = 5.5
Identities = 18/55 (32%), Positives = 26/55 (46%)
Frame = +1
Query: 7 SCIEKERHALLELKSGLVLDDTYLLPSWDTKSDDCCAWEGIGCRNQTGHVEILDL 61
+ + ++ ALL K L +L +WD D C+W G+ C N V LDL
Sbjct: 415 AAVNQQGEALLSWKRTLN-GSLEVLSNWDPVQDTPCSWYGVSC-NFKNEVVQLDL 573
>NP205681 receptor protein kinase-like protein
Length = 2946
Score = 61.6 bits (148), Expect = 8e-10
Identities = 46/138 (33%), Positives = 72/138 (51%), Gaps = 1/138 (0%)
Frame = +1
Query: 325 SNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIG 384
+N+ EGGIP FG++ SL+ LDLSS LS ++P L N+ +L L+L N + G
Sbjct: 682 NNAYEGGIPPEFGTMESLKYLDLSSCNLSGEIPPSLANM------RNLDTLFLQMNNLTG 843
Query: 385 TVP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMSM 443
T+P ++S SL ++ L N L G I + L + N+L G + S G +
Sbjct: 844 TIPSELSDMVSLMSLDLSFNGLTGEIPTRFSQLKNLTLMNFFHNNLRGSV-PSFVGELPN 1020
Query: 444 LKYLSLSSNSLALKFSEN 461
L+ L L N+ + + +N
Sbjct: 1021LETLQLWENNFSSELPQN 1074
Score = 58.5 bits (140), Expect = 6e-09
Identities = 56/184 (30%), Positives = 82/184 (44%), Gaps = 20/184 (10%)
Frame = +1
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLS--------------EDLPVMLHNLSV 365
+L I N+L G +PK +L SL+ L++S N S E L V +N +
Sbjct: 301 NLTISQNNLTGELPKELAALTSLKHLNISHNVFSGYFPGKIILPMTELEVLDVYDNNFTG 480
Query: 366 GCAKN-----SLKELYLASNQIIGTVPDM-SGFSSLENMFLYENLLNGTILKNSTFPYRL 419
+ LK L L N G++P+ S F SLE + L N L+G I K+ + L
Sbjct: 481 SLPEEFVKLEKLKYLKLDGNYFSGSIPESYSEFKSLEFLSLSTNSLSGNIPKSLSKLKTL 660
Query: 420 ANLYLDSNDLDGVITDSHFGNMSMLKYLSLSSNSLALKFSENWVPPFQLSTIYLRSCTLG 479
L L N+ FG M LKYL LSS +L+ + + L T++L+ L
Sbjct: 661 RILKLGYNNAYEGGIPPEFGTMESLKYLDLSSCNLSGEIPPSLANMRNLDTLFLQMNNLT 840
Query: 480 PSFP 483
+ P
Sbjct: 841 GTIP 852
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 58.5 bits (140), Expect = 6e-09
Identities = 40/120 (33%), Positives = 65/120 (53%), Gaps = 1/120 (0%)
Frame = +1
Query: 315 VQS*HSLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKE 374
+Q+ +L ++ N+L G + GSL SL+S+DLS+N LS ++P L +L
Sbjct: 859 LQNLDTLFLQVNALSGSLTPELGSLKSLKSMDLSNNMLSGEVPASFAEL------KNLTL 1020
Query: 375 LYLASNQIIGTVPDMSG-FSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVI 433
L L N++ G +P+ G +LE + L+EN G+I +N RL + L SN + G +
Sbjct: 1021LNLFRNKLHGAIPEFVGELPALEVLQLWENNFTGSIPQNLGNNGRLTLVDLSSNKITGTL 1200
Score = 56.6 bits (135), Expect = 2e-08
Identities = 44/132 (33%), Positives = 67/132 (50%), Gaps = 2/132 (1%)
Frame = +1
Query: 326 NSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGT 385
N+ G IP++ G+ L +DLSSNK++ LP + C N L+ L N + G
Sbjct: 1108 NNFTGSIPQNLGNNGRLTLVDLSSNKITGTLPPNM------CYGNRLQTLITLGNYLFGP 1269
Query: 386 VPDMSG-FSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMSM- 443
+PD G SL + + EN LNG+I K +L + L N L G + G+++
Sbjct: 1270 IPDSLGKCKSLNRIRMGENFLNGSIPKGLFGLPKLTQVELQDNLLTGQFPED--GSIATD 1443
Query: 444 LKYLSLSSNSLA 455
L +SLS+N L+
Sbjct: 1444 LGQISLSNNQLS 1479
Score = 40.8 bits (94), Expect = 0.001
Identities = 49/210 (23%), Positives = 87/210 (41%), Gaps = 44/210 (20%)
Frame = +1
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPV------MLHNLSVG-------- 366
L + +N P L +L LDL +N ++ +LP+ +L +L +G
Sbjct: 442 LNLSNNVFNATFPSQLNRLANLEVLDLYNNNMTGELPLSVAAMPLLRHLHLGGNFFSGQI 621
Query: 367 ----CAKNSLKELYLASNQIIGTV-PDMSGFSSLENMFL-YENLLNGTI------LKN-- 412
L+ L L+ N++ GT+ P++ SSL +++ Y N +G I L N
Sbjct: 622 PPEYGTWQHLQYLALSGNELAGTIAPELGNLSSLRELYIGYYNTYSGGIPPEIGNLSNLV 801
Query: 413 ----------STFPYRLA------NLYLDSNDLDGVITDSHFGNMSMLKYLSLSSNSLAL 456
P L L+L N L G +T G++ LK + LS+N L+
Sbjct: 802 RLDAAYCGLSGEIPAELGKLQNLDTLFLQVNALSGSLT-PELGSLKSLKSMDLSNNMLSG 978
Query: 457 KFSENWVPPFQLSTIYLRSCTLGPSFPKWI 486
+ ++ L+ + L L + P+++
Sbjct: 979 EVPASFAELKNLTLLNLFRNKLHGAIPEFV 1068
Score = 33.5 bits (75), Expect = 0.22
Identities = 21/53 (39%), Positives = 31/53 (57%), Gaps = 1/53 (1%)
Frame = +1
Query: 12 ERHALLELKSGLVLDD-TYLLPSWDTKSDDCCAWEGIGCRNQTGHVEILDLNS 63
E ALL K+ + DD T+ L SW++ S C+W G+ C ++ HV L+L S
Sbjct: 160 EYRALLSFKASSLTDDPTHALSSWNS-STPFCSWFGLTCDSRR-HVTSLNLTS 312
>TC216112 similar to UP|Q8GTD6 (Q8GTD6) Polygalacturonase inhibitor-like
protein (Fragment), partial (78%)
Length = 1175
Score = 57.8 bits (138), Expect = 1e-08
Identities = 48/135 (35%), Positives = 70/135 (51%), Gaps = 1/135 (0%)
Frame = +3
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
L +++N G IP++FGSL L LS N+LS +P + + L +L L+ N
Sbjct: 207 LDLRNNLFSGPIPRNFGSLSMLSRALLSGNRLSGAIPSSVSQI------YRLADLDLSRN 368
Query: 381 QIIGTVPDMSG-FSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFG 439
QI G +P+ G + L + L N L+G I S F +++L L N L+G I D+ FG
Sbjct: 369 QISGPIPESLGKMAVLSTLNLDMNKLSGPI-PVSLFSSGISDLNLSRNALEGNIPDA-FG 542
Query: 440 NMSMLKYLSLSSNSL 454
S L LS N+L
Sbjct: 543 VRSYFTALDLSYNNL 587
Score = 51.2 bits (121), Expect = 1e-06
Identities = 37/114 (32%), Positives = 56/114 (48%), Gaps = 1/114 (0%)
Frame = +3
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
L + N + G IP+S G + L +L+L NKLS +PV L + + + +L L+ N
Sbjct: 351 LDLSRNQISGPIPESLGKMAVLSTLNLDMNKLSGPIPVSLFS-------SGISDLNLSRN 509
Query: 381 QIIGTVPDMSGFSS-LENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVI 433
+ G +PD G S + L N L G I K+ + + +L L N L G I
Sbjct: 510 ALEGNIPDAFGVRSYFTALDLSYNNLKGAIPKSISSASYIGHLDLSHNHLCGKI 671
Score = 46.2 bits (108), Expect = 3e-05
Identities = 41/132 (31%), Positives = 59/132 (44%), Gaps = 4/132 (3%)
Frame = +3
Query: 328 LEGGIPKSFGSLCSLRSLDLSSNKLSEDLPV---MLHNLSVGCAKNSLKELYLASNQIIG 384
+ G IP+ +L LR +DL N+LS +P LH L+V L +A N I G
Sbjct: 12 ISGEIPRCITTLPFLRIVDLIGNRLSGSIPADIGRLHRLTV---------LNVADNLISG 164
Query: 385 TVP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMSM 443
T+P ++ SSL ++ L NL +G I +N FG++SM
Sbjct: 165 TIPTSLANLSSLMHLDLRNNLFSGPIPRN-------------------------FGSLSM 269
Query: 444 LKYLSLSSNSLA 455
L LS N L+
Sbjct: 270 LSRALLSGNRLS 305
Score = 32.0 bits (71), Expect = 0.65
Identities = 16/39 (41%), Positives = 23/39 (58%)
Frame = +3
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPV 358
+L + N+L+G IPKS S + LDLS N L +P+
Sbjct: 561 ALDLSYNNLKGAIPKSISSASYIGHLDLSHNHLCGKIPL 677
>TC204550 UP|Q8L3Y5 (Q8L3Y5) Receptor-like kinase RHG1, complete
Length = 2659
Score = 57.4 bits (137), Expect = 1e-08
Identities = 46/137 (33%), Positives = 67/137 (48%), Gaps = 1/137 (0%)
Frame = +2
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
+LI+ N G +P S GSL L + LS NK S +P + LS LK L +++
Sbjct: 896 NLILDHNFFTGDVPASLGSLRELNEISLSHNKFSGAIPNEIGTLS------RLKTLDISN 1057
Query: 380 NQIIGTVP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHF 438
N + G +P +S SSL + NLL+ I ++ L+ L L N G I S
Sbjct: 1058 NALNGNLPATLSNLSSLTLLNAENNLLDNQIPQSLGRLRNLSVLILSRNQFSGHIPSS-I 1234
Query: 439 GNMSMLKYLSLSSNSLA 455
N+S L+ L LS N+ +
Sbjct: 1235 ANISSLRQLDLSLNNFS 1285
Score = 42.7 bits (99), Expect = 4e-04
Identities = 24/69 (34%), Positives = 36/69 (51%)
Frame = +2
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
LI+ N G IP S ++ SLR LDLS N S ++PV ++ SL ++ N
Sbjct: 1187 LILSRNQFSGHIPSSIANISSLRQLDLSLNNFSGEIPVSFD------SQRSLNLFNVSYN 1348
Query: 381 QIIGTVPDM 389
+ G+VP +
Sbjct: 1349 SLSGSVPPL 1375
>TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, partial (92%)
Length = 3037
Score = 57.0 bits (136), Expect = 2e-08
Identities = 43/132 (32%), Positives = 68/132 (50%), Gaps = 2/132 (1%)
Frame = +3
Query: 326 NSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGT 385
N+ G IP+ G L +DLSSNKL+ LP L C+ N+L+ L N + G
Sbjct: 792 NNFTGSIPEGLGKNGRLNLVDLSSNKLTGTLPTYL------CSGNTLQTLITLGNFLFGP 953
Query: 386 VPDMSG-FSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMSM- 443
+P+ G SL + + EN LNG+I + +L + L N L G + G++++
Sbjct: 954 IPESLGSCESLTRIRMGENFLNGSIPRGLFGLPKLTQVELQDNYLSGEFPE--VGSVAVN 1127
Query: 444 LKYLSLSSNSLA 455
L ++LS+N L+
Sbjct: 1128LGQITLSNNQLS 1163
Score = 48.9 bits (115), Expect = 5e-06
Identities = 55/194 (28%), Positives = 86/194 (43%), Gaps = 26/194 (13%)
Frame = +3
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLS-SNKLSEDLPVMLHNLS--------------- 364
L + N LEG IP G+L SLR L + N + +P + NLS
Sbjct: 342 LAVSGNELEGTIPPEIGNLSSLRELYIGYYNTYTGGIPPEIGNLSELVRLDAAYCGLSGE 521
Query: 365 VGCAKNSLKE---LYLASNQIIGTV-PDMSGFSSLENMFLYENLLNGTI------LKNST 414
+ A L++ L+L N + G++ P++ SL++M L N+L+G I LKN T
Sbjct: 522 IPAALGKLQKLDTLFLQVNALSGSLTPELGNLKSLKSMDLSNNMLSGEIPARFGELKNIT 701
Query: 415 FPYRLANLYLDSNDLDGVITDSHFGNMSMLKYLSLSSNSLALKFSENWVPPFQLSTIYLR 474
L NL+ N L G I + G + L+ + L N+ E +L+ + L
Sbjct: 702 ----LLNLF--RNKLHGAIPE-FIGELPALEVVQLWENNFTGSIPEGLGKNGRLNLVDLS 860
Query: 475 SCTLGPSFPKWIRS 488
S L + P ++ S
Sbjct: 861 SNKLTGTLPTYLCS 902
Score = 39.7 bits (91), Expect = 0.003
Identities = 31/107 (28%), Positives = 47/107 (42%), Gaps = 1/107 (0%)
Frame = +3
Query: 326 NSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGT 385
N G I L LDLS N+LS D+P + + + L L L+ N ++G
Sbjct: 1296 NKFSGPIVPEISQCKLLTFLDLSRNELSGDIPNEITGMRI------LNYLNLSRNHLVGG 1457
Query: 386 VP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDG 431
+P +S SL ++ N L+G + F Y +L + DL G
Sbjct: 1458 IPSSISSMQSLTSVDFSYNNLSGLVPGTGQFSYFNYTSFLGNPDLCG 1598
>BQ081742
Length = 407
Score = 57.0 bits (136), Expect = 2e-08
Identities = 41/115 (35%), Positives = 62/115 (53%), Gaps = 1/115 (0%)
Frame = +2
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
+L + +N+L G IPK GSL L L+LS+NK +E +P SL++L L
Sbjct: 35 NLELAANNLGGPIPKQVGSLHKLLHLNLSNNKFTESIPSFNQ-------LQSLQDLDLGR 193
Query: 380 NQIIGTVP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVI 433
N + G +P +++ LE + L N L+GTI F LAN+ + +N L+G I
Sbjct: 194 NLLNGKIPAELATLQRLETLNLSHNNLSGTI---PDFKNSLANVDISNNQLEGSI 349
Score = 41.6 bits (96), Expect = 8e-04
Identities = 26/78 (33%), Positives = 40/78 (50%)
Frame = +2
Query: 315 VQS*HSLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKE 374
+QS L + N L G IP +L L +L+LS N LS +P KNSL
Sbjct: 161 LQSLQDLDLGRNLLNGKIPAELATLQRLETLNLSHNNLSGTIPDF---------KNSLAN 313
Query: 375 LYLASNQIIGTVPDMSGF 392
+ +++NQ+ G++P + F
Sbjct: 314 VDISNNQLEGSIPSIPAF 367
>TC205792 similar to UP|Q93X72 (Q93X72) Leucine-rich repeat resistance
protein-like protein, partial (93%)
Length = 1540
Score = 56.2 bits (134), Expect = 3e-08
Identities = 42/134 (31%), Positives = 65/134 (48%), Gaps = 1/134 (0%)
Frame = +1
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
L + N L G IP G+L +LR LD +N L + ++ GC +L+ LYL +N
Sbjct: 799 LYLHENRLAGRIPPELGTLQNLRHLDAGNNHLVGTIRELIR--IEGCFP-ALRNLYLNNN 969
Query: 381 QIIGTVP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFG 439
G +P ++ +SLE ++L N ++G I +L LYLD N G I + F
Sbjct: 970 YFTGGIPAQLANLTSLEILYLSYNKMSGVIPSTVAHIPKLTYLYLDHNQFSGRIPEP-FY 1146
Query: 440 NMSMLKYLSLSSNS 453
LK + + N+
Sbjct: 1147KHPFLKEMYIEGNA 1188
Score = 45.8 bits (107), Expect = 4e-05
Identities = 40/129 (31%), Positives = 58/129 (44%), Gaps = 1/129 (0%)
Frame = +1
Query: 327 SLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGTV 386
S+ G P + SL L LDL +NKL+ +P + L LK L L N++ +
Sbjct: 529 SIVGPFPTAVTSLLDLTRLDLHNNKLTGPIPPQIGRL------KRLKILNLRWNKLQDAI 690
Query: 387 -PDMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMSMLK 445
P++ SL +++L N G I K L LYL N L G I G + L+
Sbjct: 691 PPEIGELKSLTHLYLSFNNFKGEIPKELANLPDLRYLYLHENRLAGRI-PPELGTLQNLR 867
Query: 446 YLSLSSNSL 454
+L +N L
Sbjct: 868 HLDAGNNHL 894
Score = 40.0 bits (92), Expect = 0.002
Identities = 29/97 (29%), Positives = 44/97 (44%)
Frame = +1
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
+L + +N GGIP +L SL L LS NK+S +P + ++ L LYL
Sbjct: 949 NLYLNNNYFTGGIPAQLANLTSLEILYLSYNKMSGVIPSTVAHIP------KLTYLYLDH 1110
Query: 380 NQIIGTVPDMSGFSSLENMFLYENLLNGTILKNSTFP 416
NQ G +P+ ++ FL E + G + P
Sbjct: 1111 NQFSGRIPE----PFYKHPFLKEMYIEGNAFRPGVNP 1209
>TC230707 similar to UP|Q12937 (Q12937) BS-84, partial (4%)
Length = 990
Score = 55.8 bits (133), Expect = 4e-08
Identities = 44/135 (32%), Positives = 76/135 (55%), Gaps = 1/135 (0%)
Frame = +3
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
+L ++SNS G IP S +L SL SLDL+ N LS LP +++L+ +L+ L L+
Sbjct: 99 TLTLRSNSFSGTIPPSITTLKSLLSLDLAHNSLSGYLPNSMNSLT------TLRRLDLSF 260
Query: 380 NQIIGTVPDMSGFSSLENMFLYENLLNGTILKNS-TFPYRLANLYLDSNDLDGVITDSHF 438
N++ G++P + S+L + + N L+G + K S +L + L N L G + +S F
Sbjct: 261 NKLTGSIPKLP--SNLLELAIKANSLSGPLQKQSFEGMNQLEVVELSENALTGTV-ESWF 431
Query: 439 GNMSMLKYLSLSSNS 453
+ L+ + L++N+
Sbjct: 432 FLLPSLQQVDLANNT 476
Score = 53.5 bits (127), Expect = 2e-07
Identities = 47/144 (32%), Positives = 64/144 (43%), Gaps = 2/144 (1%)
Frame = +3
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
SL + NSL G +P S SL +LR LDLS NKL+ +P + N L EL + +
Sbjct: 171 SLDLAHNSLSGYLPNSMNSLTTLRRLDLSFNKLTGSIPKLPSN---------LLELAIKA 323
Query: 380 NQIIGTVPDMS--GFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSH 437
N + G + S G + LE + L EN L GT+ L + L +N GV
Sbjct: 324 NSLSGPLQKQSFEGMNQLEVVELSENALTGTVESWFFLLPSLQQVDLANNTFTGVQISRP 503
Query: 438 FGNMSMLKYLSLSSNSLALKFSEN 461
S +SN +AL N
Sbjct: 504 LAARGGSSSSSGNSNLVALNLGFN 575
>TC213579 weakly similar to UP|O50024 (O50024) Hcr9-4E, partial (13%)
Length = 1007
Score = 54.7 bits (130), Expect = 9e-08
Identities = 33/83 (39%), Positives = 45/83 (53%)
Frame = +1
Query: 315 VQS*HSLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKE 374
+ S L + N L G IP S G+L +L SLDLSSN L+ +P L NL N L+
Sbjct: 295 LHSLRGLNLSHNRLRGPIPNSMGNLTNLESLDLSSNMLTGRIPTGLTNL------NFLEV 456
Query: 375 LYLASNQIIGTVPDMSGFSSLEN 397
L L++N +G +P FS+ N
Sbjct: 457 LNLSNNHFVGEIPQGKQFSTFSN 525
>TC227239 similar to UP|Q6NQP4 (Q6NQP4) At3g43740, partial (79%)
Length = 1051
Score = 53.5 bits (127), Expect = 2e-07
Identities = 38/113 (33%), Positives = 57/113 (49%)
Frame = +3
Query: 299 IYRVTIWQAVSMVS*SVQS*HSLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPV 358
+YR I + +++S S+ + N LEG IPKSFG L SL+ L L++NKL+ +P
Sbjct: 375 LYRNEITGKIPKELGNLKSLISMDLYDNKLEGKIPKSFGKLKSLKFLRLNNNKLTGSIPR 554
Query: 359 MLHNLSVGCAKNSLKELYLASNQIIGTVPDMSGFSSLENMFLYENLLNGTILK 411
L L+ LK +++N + GT+P F S N +G LK
Sbjct: 555 ELTRLT------DLKIFDVSNNDLCGTIPVEGNFESFPMESFKNNRFSGPELK 695
Score = 28.1 bits (61), Expect = 9.3
Identities = 19/52 (36%), Positives = 26/52 (49%)
Frame = +3
Query: 12 ERHALLELKSGLVLDDTYLLPSWDTKSDDCCAWEGIGCRNQTGHVEILDLNS 63
E +AL L+S L D +L SWD + C W + C + HV LDL +
Sbjct: 162 EGNALHALRSRLS-DPNNMLQSWDPTLVNPCTWFHVTC-DSNNHVIRLDLGN 311
>NP206608 unknown
Length = 477
Score = 53.1 bits (126), Expect = 3e-07
Identities = 29/63 (46%), Positives = 38/63 (60%)
Frame = +1
Query: 7 SCIEKERHALLELKSGLVLDDTYLLPSWDTKSDDCCAWEGIGCRNQTGHVEILDLNSDQF 66
+C EKER+ALL K GL D + L SW KSD CC W G+ C N TG V ++L++
Sbjct: 97 TCSEKERNALLSFKHGLA-DPSNRLSSWSDKSD-CCTWPGVHC-NNTGKVMEINLDTPAG 267
Query: 67 GPF 69
P+
Sbjct: 268 SPY 276
>TC213089 similar to PIR|T05606|T05606 protein kinase homolog F9D16.210 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(21%)
Length = 740
Score = 52.8 bits (125), Expect = 4e-07
Identities = 38/98 (38%), Positives = 55/98 (55%), Gaps = 7/98 (7%)
Frame = +2
Query: 371 SLKELYLASNQIIGTVPDMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLD 430
+L LYL N I G +PD S + +L + L +N NGTI + + +LA L L +N L
Sbjct: 26 NLSFLYLQFNNISGPLPDFSAWKNLTVVNLSDNHFNGTIPSSLSKLTQLAGLNLANNTLS 205
Query: 431 GVITDSHFGNMSMLKYLSLSSNSL-------ALKFSEN 461
G I D N+S L+ L+LS+N+L L+FSE+
Sbjct: 206 GEIPDL---NLSRLQVLNLSNNNLQGSVPKSLLRFSES 310
>TC230883 UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, complete
Length = 3053
Score = 52.8 bits (125), Expect = 4e-07
Identities = 37/126 (29%), Positives = 63/126 (49%), Gaps = 19/126 (15%)
Frame = +1
Query: 327 SLEGGIPKSFGSLCSLRSLDLSSNKLSEDLP-----------VMLHNLSVGCA------- 368
SL G +P F SL+ L LS N L+ +LP + L+N + G +
Sbjct: 421 SLTGPLPDIFDKFPSLQHLRLSYNNLTGNLPSSFSAANNLETLWLNNQAAGLSGTLLVLS 600
Query: 369 -KNSLKELYLASNQIIGTVPDMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSN 427
++L + +L NQ G++PD+S ++L ++ L +N L G + + T L + LD+N
Sbjct: 601 NMSALNQSWLNKNQFTGSIPDLSQCTALSDLQLRDNQLTGVVPASLTSLPSLKKVSLDNN 780
Query: 428 DLDGVI 433
+L G +
Sbjct: 781 ELQGPV 798
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.338 0.146 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,724,895
Number of Sequences: 63676
Number of extensions: 386177
Number of successful extensions: 3502
Number of sequences better than 10.0: 173
Number of HSP's better than 10.0 without gapping: 3298
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3459
length of query: 491
length of database: 12,639,632
effective HSP length: 101
effective length of query: 390
effective length of database: 6,208,356
effective search space: 2421258840
effective search space used: 2421258840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC136507.3


