

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
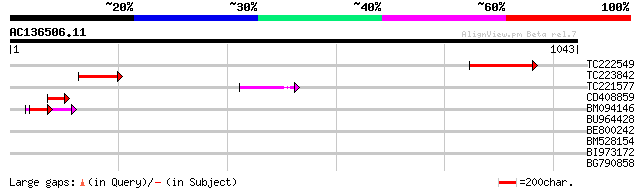
Query= AC136506.11 - phase: 0 /pseudo
(1043 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC222549 weakly similar to UP|Q6YXW5 (Q6YXW5) Cyclin-like protei... 132 6e-31
TC223842 weakly similar to GB|AAQ89612.1|37201994|BT010590 At5g2... 78 2e-14
TC221577 similar to UP|Q6YXW5 (Q6YXW5) Cyclin-like protein, part... 55 2e-07
CD408859 weakly similar to GP|14194169|gb| At1g05960/T21E18_20 {... 55 2e-07
BM094146 similar to GP|14194169|gb| At1g05960/T21E18_20 {Arabido... 54 4e-07
BU964428 weakly similar to GP|14194169|gb| At1g05960/T21E18_20 {... 41 0.002
BE800242 37 0.059
BM528154 35 0.17
BI973172 similar to GP|14194169|gb| At1g05960/T21E18_20 {Arabido... 34 0.38
BG790858 weakly similar to GP|14194169|gb| At1g05960/T21E18_20 {... 32 1.5
>TC222549 weakly similar to UP|Q6YXW5 (Q6YXW5) Cyclin-like protein, partial
(3%)
Length = 388
Score = 132 bits (333), Expect = 6e-31
Identities = 79/127 (62%), Positives = 86/127 (67%), Gaps = 2/127 (1%)
Frame = +2
Query: 846 GCPVIS*YNR--IPVWTKG**TF*YG*CSIIYY***HSSFWFGKPN*Y*CATTAFRKS*P 903
G PVI NR +PVWTK *T *YG* IIYY* *HS+ WFG+P+*+* TA KS P
Sbjct: 8 GSPVICRDNRKYVPVWTKRR*TS*YG*YFIIYY*R*HSTLWFGEPS*F*FNATALTKSKP 187
Query: 904 DNC**YFGFSCGDNTSCRKDFCFNTF*HAI*RNGSPL*EPPRRKATEDFHIHGCPIITCQ 963
C**+FGFS GDNTS R DF FNT *HA+ NGSPL* PPR KA ED H HG P
Sbjct: 188 AEC**HFGFSFGDNTSSRTDFYFNTV*HALQGNGSPL*GPPRGKAAEDVHFHGYPSNARI 367
Query: 964 FIQDSSS 970
FI DS S
Sbjct: 368 FI*DSCS 388
>TC223842 weakly similar to GB|AAQ89612.1|37201994|BT010590 At5g26850
{Arabidopsis thaliana;} , partial (8%)
Length = 911
Score = 77.8 bits (190), Expect = 2e-14
Identities = 37/81 (45%), Positives = 53/81 (64%)
Frame = +2
Query: 127 SRQDEVQILGCQTLFDFVNNQRDGTYMFNLDSFILKLCHLAQQVGDDGKVEHLRASGLQV 186
S+ + +Q LGCQ L F+ Q D TY N++ + K+C L+++ G+ + L AS LQ
Sbjct: 38 SKDETIQTLGCQCLSKFIYCQVDATYTHNIEKLVPKVCMLSREHGEACEKRCLSASSLQC 217
Query: 187 LSSMVWFMGEFTHISVEFDNV 207
LS+MVWFM EF+HI V+FD V
Sbjct: 218 LSAMVWFMAEFSHIFVDFDEV 280
>TC221577 similar to UP|Q6YXW5 (Q6YXW5) Cyclin-like protein, partial (13%)
Length = 775
Score = 54.7 bits (130), Expect = 2e-07
Identities = 35/111 (31%), Positives = 60/111 (53%)
Frame = +2
Query: 423 CRSCSRYHGRVVRKHV*YYSYGKNLDCCSL*NFSNSCIYTEFVVSEQGIP*GIISSVTPG 482
CR+ S + G VR+++ Y + +N + C L N + +++ ++S++G P* +SS+ PG
Sbjct: 275 CRTHS*FDGCGVREYLKYSHHSRNNNLCCLSNCKVNHVHS*CIISQKGFP*CFVSSIAPG 454
Query: 483 NGPCRP*NQGGCTPHIFSCSCAIVSLSSTFIFKSPLGKGNRYSKNALEKCL 533
+G P*N T H+F + AI S F SP+ N+Y ++ L
Sbjct: 455 DGSS*P*NTSWSTQHLFFSTYAI----SIF---SPVRPKNKYFSEGTKRKL 586
>CD408859 weakly similar to GP|14194169|gb| At1g05960/T21E18_20 {Arabidopsis
thaliana}, partial (4%)
Length = 464
Score = 54.7 bits (130), Expect = 2e-07
Identities = 22/40 (55%), Positives = 31/40 (77%)
Frame = -3
Query: 70 KITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQM 109
+I LE RCYKEL+ EN+ + K+V+CIY+K L SC++QM
Sbjct: 330 RIVQALEPRCYKELQNENFISTKIVMCIYKKFLFSCKEQM 211
>BM094146 similar to GP|14194169|gb| At1g05960/T21E18_20 {Arabidopsis
thaliana}, partial (6%)
Length = 398
Score = 53.9 bits (128), Expect = 4e-07
Identities = 24/42 (57%), Positives = 28/42 (66%)
Frame = +3
Query: 37 LADIFPRTPEEEPNDRKISKLCEYASKNPLRVPKITSYLEQR 78
L FPR E PNDR I KLC+Y ++NPL +PKI LEQR
Sbjct: 27 LLTFFPRNQVEVPNDRSIGKLCDYTARNPLCIPKIVQALEQR 152
Score = 45.4 bits (106), Expect = 1e-04
Identities = 30/95 (31%), Positives = 47/95 (48%), Gaps = 1/95 (1%)
Frame = +2
Query: 29 PIKRYKKLLADIFPRTPEEEPNDRKISKLC-EYASKNPLRVPKITSYLEQRCYKELRTEN 87
P+KRYKKL+ADIFP P K K+ Y K S++ + +
Sbjct: 2 PVKRYKKLIADIFPS*PGGSAK**KHWKIM*LYCQK---------SFMHPKDCASP*AKV 154
Query: 88 YQAVKVVICIYRKLLVSCRDQMPLFASSLLSIIQI 122
+ K+V+CIY+K L SC++QM + + ++ I
Sbjct: 155 ITSTKIVMCIYKKFLFSCKEQM*VHIYLIFKLVYI 259
>BU964428 weakly similar to GP|14194169|gb| At1g05960/T21E18_20 {Arabidopsis
thaliana}, partial (10%)
Length = 253
Score = 41.2 bits (95), Expect = 0.002
Identities = 22/52 (42%), Positives = 28/52 (53%)
Frame = +1
Query: 259 CYEPWILVKSLYTKYG*VSQGRYNCTTCPGILVPLF**YQSLVSRTRACPIC 310
C +ILVK L YG + +G YN T CP LF* +S+V R+C C
Sbjct: 25 C*RSYILVKGLLVSYGQIGKGSYNITACPRASFSLF*Y*KSMVF*KRSC*SC 180
>BE800242
Length = 247
Score = 36.6 bits (83), Expect = 0.059
Identities = 20/62 (32%), Positives = 35/62 (56%)
Frame = +2
Query: 681 EFFS*GSNSKFSVGIFLEEHFSQ*KW*TLLLSFFS*TASITPQVSLYTSNIHDCLHIQGL 740
EFF *G++S+FS F E+F + A+I+ +++LY N++DC+ + +
Sbjct: 44 EFFL*GTSSEFSACFFFMEYFP*RRA----------IATISSEITLYAGNVNDCVFFKIV 193
Query: 741 QH 742
QH
Sbjct: 194 QH 199
>BM528154
Length = 428
Score = 35.0 bits (79), Expect = 0.17
Identities = 16/31 (51%), Positives = 21/31 (67%)
Frame = +2
Query: 184 LQVLSSMVWFMGEFTHISVEFDNVSLASLVN 214
LQ LS MV FMGE +H+S++FD + L N
Sbjct: 2 LQALSHMVQFMGEHSHLSMDFDKIISVILEN 94
Score = 31.2 bits (69), Expect = 2.5
Identities = 18/60 (30%), Positives = 29/60 (48%)
Frame = +1
Query: 257 GRCYEPWILVKSLYTKYG*VSQGRYNCTTCPGILVPLF**YQSLVSRTRACPICLARYAV 316
G C +LVK+L Y + +G Y+C CP F* + +V +C +C +A+
Sbjct: 211 GCC*RSCLLVKALLV*YCQIGKGSYHCAACPQTTFS*F*F*KPMVF*KGSCFMCFDVFAI 390
>BI973172 similar to GP|14194169|gb| At1g05960/T21E18_20 {Arabidopsis
thaliana}, partial (5%)
Length = 420
Score = 33.9 bits (76), Expect = 0.38
Identities = 16/45 (35%), Positives = 27/45 (59%)
Frame = +1
Query: 440 YYSYGKNLDCCSL*NFSNSCIYTEFVVSEQGIP*GIISSVTPGNG 484
YY + K + C L N + + + V+S+ G P* ++SS+ PG+G
Sbjct: 16 YYHHSKINNHCCLSNSKINHFHPQCVISQ*GFP*CLVSSIAPGHG 150
>BG790858 weakly similar to GP|14194169|gb| At1g05960/T21E18_20 {Arabidopsis
thaliana}, partial (11%)
Length = 417
Score = 32.0 bits (71), Expect = 1.5
Identities = 19/51 (37%), Positives = 25/51 (48%)
Frame = +2
Query: 266 VKSLYTKYG*VSQGRYNCTTCPGILVPLF**YQSLVSRTRACPICLARYAV 316
VK L Y + +G YNC CP F** + LV +C +C +AV
Sbjct: 2 VKGLLV*YCQIGKGSYNCAACPRTAFS*F***KPLVF*KGSCFLCFDVFAV 154
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.361 0.161 0.605
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 52,937,153
Number of Sequences: 63676
Number of extensions: 837106
Number of successful extensions: 8751
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 4776
Number of HSP's successfully gapped in prelim test: 337
Number of HSP's that attempted gapping in prelim test: 3830
Number of HSP's gapped (non-prelim): 5433
length of query: 1043
length of database: 12,639,632
effective HSP length: 107
effective length of query: 936
effective length of database: 5,826,300
effective search space: 5453416800
effective search space used: 5453416800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC136506.11


