

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136504.6 - phase: 0 /pseudo
(417 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
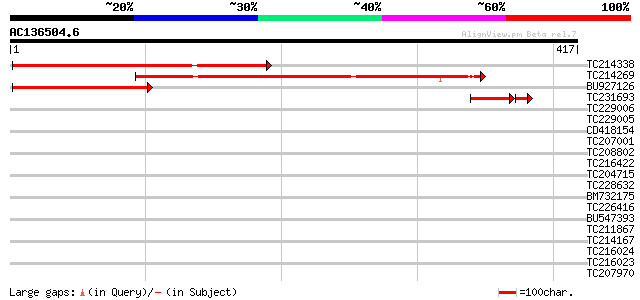
Score E
Sequences producing significant alignments: (bits) Value
TC214338 similar to UP|Q9ASR5 (Q9ASR5) AT4g35920/F4B14_190, part... 330 7e-91
TC214269 similar to UP|Q9ASR5 (Q9ASR5) AT4g35920/F4B14_190, part... 296 1e-80
BU927126 similar to GP|13605847|gb AT4g35920/F4B14_190 {Arabidop... 194 8e-50
TC231693 homologue to UP|Q9ASR5 (Q9ASR5) AT4g35920/F4B14_190, pa... 43 8e-06
TC229006 39 0.006
TC229005 37 0.022
CD418154 weakly similar to GP|13605847|gb| AT4g35920/F4B14_190 {... 32 0.059
TC207001 similar to PIR|F86340|F86340 protein F2D10.34 [imported... 32 0.69
TC208802 weakly similar to GB|AAM78037.1|21928017|AY125527 AT3g4... 30 1.5
TC216422 30 2.0
TC204715 similar to UP|Q8H0X0 (Q8H0X0) Expressed protein, partia... 30 2.0
TC228632 similar to UP|Q9ZTQ9 (Q9ZTQ9) MADS box protein 26, part... 30 2.0
BM732175 similar to GP|15022157|gb MADS box protein-like protein... 30 2.6
TC226416 homologue to UP|MDHP_MEDSA (O48902) Malate dehydrogenas... 29 3.4
BU547393 weakly similar to GP|4164408|emb amino acid carrier {Ri... 28 5.9
TC211867 weakly similar to UP|PNX1_HUMAN (P55347) Homeobox prote... 28 5.9
TC214167 similar to UP|Q6NLW5 (Q6NLW5) At5g48720, partial (30%) 28 5.9
TC216024 similar to GB|AAM78036.1|21928015|AY125526 At2g01100/F2... 28 5.9
TC216023 similar to GB|AAM78036.1|21928015|AY125526 At2g01100/F2... 28 5.9
TC207970 weakly similar to UP|Q94A73 (Q94A73) AT5g66560/K1F13_23... 28 5.9
>TC214338 similar to UP|Q9ASR5 (Q9ASR5) AT4g35920/F4B14_190, partial (42%)
Length = 889
Score = 330 bits (846), Expect = 7e-91
Identities = 167/190 (87%), Positives = 176/190 (91%)
Frame = +2
Query: 3 SSWEQFGEIANVAQLTGLDAVKLIGMIVKAANTARMHKKNCRQFAQHLKLIGNLLDQLKI 62
+SW+Q GE+ANVAQLTG+DAV+LIGMIV+AA+TARMHKKNCRQFAQHLKLIGNLL+QLKI
Sbjct: 329 ASWDQMGELANVAQLTGVDAVRLIGMIVRAASTARMHKKNCRQFAQHLKLIGNLLEQLKI 508
Query: 63 SELKKYPETREPLEQLEDALRRSYILVNSCQDRSYLYLLAMGWNIVYQFRKAQNEIDRYL 122
SELKKYPETREPLEQLEDALRRSYILVNSCQDRSYLYLLAMGWNIVYQFRKAQNEIDRYL
Sbjct: 509 SELKKYPETREPLEQLEDALRRSYILVNSCQDRSYLYLLAMGWNIVYQFRKAQNEIDRYL 688
Query: 123 RLVPLITLVDNARATRVRERLEVIEKDQCAYTLDDEDQKVQTVFMKPEHDKEDTVVLKKT 182
LVPLITLVDNA RVRERLEVIE DQ YTLDDEDQK QTV KPE DK+DT VLKKT
Sbjct: 689 PLVPLITLVDNA---RVRERLEVIEMDQREYTLDDEDQKAQTVIFKPEPDKDDTAVLKKT 859
Query: 183 LSCSYPNFSF 192
LSCSYPN SF
Sbjct: 860 LSCSYPNCSF 889
>TC214269 similar to UP|Q9ASR5 (Q9ASR5) AT4g35920/F4B14_190, partial (40%)
Length = 1749
Score = 296 bits (758), Expect = 1e-80
Identities = 165/260 (63%), Positives = 192/260 (73%), Gaps = 2/260 (0%)
Frame = -3
Query: 93 QDRSYLYLLAMGWNIVYQFRKAQNEIDRYLRLVPLITLVDNARATRVRERLEVIEKDQCA 152
+DRSYLYLLAMGWNIVYQFRKAQNEIDRYLRLVPLITLVDN+R VRERLEVIE DQ
Sbjct: 961 RDRSYLYLLAMGWNIVYQFRKAQNEIDRYLRLVPLITLVDNSR---VRERLEVIEMDQRE 791
Query: 153 YTLDDEDQKVQTVFMKPEHDKEDTVVLKKTLSCSYPNFSFTEALKKENEKLQLELHHSQA 212
YTLDDEDQK QTV KPE DK+DT VLKKTLSCSYPN SFTEALKKENEKL+LEL SQA
Sbjct: 790 YTLDDEDQKAQTVIFKPEPDKDDTAVLKKTLSCSYPNCSFTEALKKENEKLKLELQRSQA 611
Query: 213 NMDINQCEFIQRLLDVTNFAAYSLPEKLSPEDNYQIVEYSNNSHSDANGSKGHSSNEKHH 272
N+D+NQCE IQRLLDVT AAYS+P K SPE +++ EY ++SDAN K HSS+EK+H
Sbjct: 610 NLDMNQCEVIQRLLDVTEVAAYSVPAKCSPEKSHKKEEY---NYSDANSDKDHSSDEKYH 440
Query: 273 KKKPKFQKRIRCQLEVHISRKIGILIYLLVVLNLIFIFSFAGI--KTFFYPCGTFSKIAT 330
K K + ++ G +L+ S + KTFFYPCGTFSKIA+
Sbjct: 439 AKIDKHSPSRYSVAQKDLASTGGSYQQEDWHTDLLACCSEPSLCMKTFFYPCGTFSKIAS 260
Query: 331 VATNRPIFCRSMQ*LYCIFV 350
VA NRPI C+ + ++C+ +
Sbjct: 259 VARNRPI-CK-LAIMHCVSI 206
>BU927126 similar to GP|13605847|gb AT4g35920/F4B14_190 {Arabidopsis
thaliana}, partial (23%)
Length = 453
Score = 194 bits (492), Expect = 8e-50
Identities = 93/103 (90%), Positives = 101/103 (97%)
Frame = +3
Query: 3 SSWEQFGEIANVAQLTGLDAVKLIGMIVKAANTARMHKKNCRQFAQHLKLIGNLLDQLKI 62
+SW+Q GE+ANVAQLTG+DAV+LIGM V+AA+TARMHKKNCRQFAQHLKLIGNLL+QLKI
Sbjct: 144 ASWDQMGELANVAQLTGVDAVRLIGMFVRAASTARMHKKNCRQFAQHLKLIGNLLEQLKI 323
Query: 63 SELKKYPETREPLEQLEDALRRSYILVNSCQDRSYLYLLAMGW 105
SELKKYPETREPLEQLEDALRRSYILVNSCQDRSYLYLLAMGW
Sbjct: 324 SELKKYPETREPLEQLEDALRRSYILVNSCQDRSYLYLLAMGW 452
>TC231693 homologue to UP|Q9ASR5 (Q9ASR5) AT4g35920/F4B14_190, partial (14%)
Length = 556
Score = 42.7 bits (99), Expect(2) = 8e-06
Identities = 24/32 (75%), Positives = 25/32 (78%)
Frame = +1
Query: 340 RSMQ*LYCIFVGFILLLLYLLHKEEA*EDDEH 371
RSM *L IFV F+LLLLYLL KEEA ED EH
Sbjct: 37 RSM**LDGIFVNFVLLLLYLLRKEEASEDVEH 132
Score = 24.6 bits (52), Expect(2) = 8e-06
Identities = 9/12 (75%), Positives = 11/12 (91%)
Frame = +2
Query: 373 EWREVQIRGVYG 384
EWREV+IRG+ G
Sbjct: 197 EWREVEIRGLTG 232
>TC229006
Length = 991
Score = 38.5 bits (88), Expect = 0.006
Identities = 23/85 (27%), Positives = 44/85 (51%)
Frame = +2
Query: 145 VIEKDQCAYTLDDEDQKVQTVFMKPEHDKEDTVVLKKTLSCSYPNFSFTEALKKENEKLQ 204
V+ + T+D++D+K+Q + + H K ++ L+ SF + +++ NEKL
Sbjct: 410 VVSPREARQTMDEKDKKIQELTAELRHKKRLCATYQEQLT------SFMKIVEEHNEKLS 571
Query: 205 LELHHSQANMDINQCEFIQRLLDVT 229
++HH N + + E I+ LL T
Sbjct: 572 AKIHHVVNN--LKEFESIEELLHQT 640
>TC229005
Length = 679
Score = 36.6 bits (83), Expect = 0.022
Identities = 25/87 (28%), Positives = 45/87 (50%), Gaps = 2/87 (2%)
Frame = +1
Query: 145 VIEKDQCAYTLDDEDQKVQTVFMKPEHDKEDTVVLKKTLSCSYPN--FSFTEALKKENEK 202
V+ + T+D++D+K+Q + + + LKK L +Y SF + +++ NEK
Sbjct: 103 VVSPREARQTMDEKDKKIQELTAE--------LRLKKRLCATYQEQLSSFMKIVEERNEK 258
Query: 203 LQLELHHSQANMDINQCEFIQRLLDVT 229
L ++HH N + + E I+ LL T
Sbjct: 259 LSAKIHHVVNN--LKEFESIEELLHQT 333
>CD418154 weakly similar to GP|13605847|gb| AT4g35920/F4B14_190 {Arabidopsis
thaliana}, partial (19%)
Length = 595
Score = 31.6 bits (70), Expect(2) = 0.059
Identities = 13/19 (68%), Positives = 15/19 (78%)
Frame = -3
Query: 315 IKTFFYPCGTFSKIATVAT 333
+KT F+PCGTFS IA V T
Sbjct: 518 LKTCFFPCGTFSWIANVVT 462
Score = 22.3 bits (46), Expect(2) = 0.059
Identities = 9/11 (81%), Positives = 11/11 (99%)
Frame = -2
Query: 354 LLLLYLLHKEE 364
LLLL+LLHK+E
Sbjct: 399 LLLLFLLHKKE 367
>TC207001 similar to PIR|F86340|F86340 protein F2D10.34 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(54%)
Length = 1074
Score = 31.6 bits (70), Expect = 0.69
Identities = 29/104 (27%), Positives = 42/104 (39%), Gaps = 2/104 (1%)
Frame = +2
Query: 237 PEKLSPEDNYQIVEYSN--NSHSDANGSKGHSSNEKHHKKKPKFQKRIRCQLEVHISRKI 294
PE S EDN SN N++SD+ G+ + + E HH H++
Sbjct: 698 PELKSREDNNVASNSSNNINNNSDSGGNNSNCNIEHHHSH--------------HVNSGF 835
Query: 295 GILIYLLVVLNLIFIFSFAGIKTFFYPCGTFSKIATVATNRPIF 338
L+ ++ F F+F FYP F KI A P+F
Sbjct: 836 LP*T**LLFISFFFFFNFVP----FYPFKLFKKIRNWAFFSPLF 955
>TC208802 weakly similar to GB|AAM78037.1|21928017|AY125527
AT3g46450/F18L15_170 {Arabidopsis thaliana;} , partial
(55%)
Length = 1539
Score = 30.4 bits (67), Expect = 1.5
Identities = 16/75 (21%), Positives = 36/75 (47%), Gaps = 10/75 (13%)
Frame = +3
Query: 193 TEALKKENEKLQLELHHSQANMDINQCEFIQRLLDVTNFAAY----------SLPEKLSP 242
++ + + + EL ++AN++ + CE +L+ N+ SLPE+++
Sbjct: 753 SDCVASHDSEASAELSPAEANLNTSHCEIDPEVLEYENWLTLLNQELENQGISLPERIND 932
Query: 243 EDNYQIVEYSNNSHS 257
++ ++ SNN S
Sbjct: 933 DELHRFYTASNNDSS 977
>TC216422
Length = 899
Score = 30.0 bits (66), Expect = 2.0
Identities = 17/52 (32%), Positives = 23/52 (43%)
Frame = -1
Query: 180 KKTLSCSYPNFSFTEALKKENEKLQLELHHSQANMDINQCEFIQRLLDVTNF 231
K + S NF F N K+ + ++ SQ N I C F L VTN+
Sbjct: 779 KSNI*TSIRNFKFLLIHYINNRKISIAINWSQENSPIIVCHFFSSCLGVTNW 624
>TC204715 similar to UP|Q8H0X0 (Q8H0X0) Expressed protein, partial (76%)
Length = 1150
Score = 30.0 bits (66), Expect = 2.0
Identities = 12/47 (25%), Positives = 26/47 (54%)
Frame = +3
Query: 349 FVGFILLLLYLLHKEEA*EDDEHSEWREVQIRGVYGMTSSSIFLKHC 395
F+G +++LLY++ + S W +Q G++ + +I++ HC
Sbjct: 789 FIGVLIILLYVV--------GDLSYWTSLQFEGLFHVIGENIYVGHC 905
>TC228632 similar to UP|Q9ZTQ9 (Q9ZTQ9) MADS box protein 26, partial (82%)
Length = 880
Score = 30.0 bits (66), Expect = 2.0
Identities = 18/48 (37%), Positives = 26/48 (53%), Gaps = 2/48 (4%)
Frame = +1
Query: 194 EALKKENEKLQLELHHSQANMDINQCEF--IQRLLDVTNFAAYSLPEK 239
E LKKEN+ +Q+EL H + + DIN + + L D S+ EK
Sbjct: 328 ERLKKENDSMQIELRHLKGD-DINSLNYKELMALEDALETGLVSVREK 468
>BM732175 similar to GP|15022157|gb MADS box protein-like protein NGL9
{Medicago sativa}, partial (60%)
Length = 421
Score = 29.6 bits (65), Expect = 2.6
Identities = 18/48 (37%), Positives = 25/48 (51%), Gaps = 2/48 (4%)
Frame = +2
Query: 194 EALKKENEKLQLELHHSQANMDINQCEF--IQRLLDVTNFAAYSLPEK 239
E LKKEN+ +Q+EL H + DIN + + L D S+ EK
Sbjct: 245 ERLKKENDSMQIELRHLKGE-DINSLNYKELMALEDALETGLVSVREK 385
>TC226416 homologue to UP|MDHP_MEDSA (O48902) Malate dehydrogenase [NADP],
chloroplast precursor (NADP-MDH) , partial (96%)
Length = 1767
Score = 29.3 bits (64), Expect = 3.4
Identities = 25/85 (29%), Positives = 43/85 (50%)
Frame = -1
Query: 121 YLRLVPLITLVDNARATRVRERLEVIEKDQCAYTLDDEDQKVQTVFMKPEHDKEDTVVLK 180
+ +L P + LV R + + E EV +K C+ + +E ++ K ED V+ K
Sbjct: 363 FSQLAPALHLVQQNRKS*LVEFYEVAQK--CSGDV*EE-----SLIEKRAEMSEDEVLSK 205
Query: 181 KTLSCSYPNFSFTEALKKENEKLQL 205
+ S+P TE+LKK ++K+ L
Sbjct: 204 WGSTVSHPWQHSTESLKKRSKKIAL 130
>BU547393 weakly similar to GP|4164408|emb amino acid carrier {Ricinus
communis}, partial (33%)
Length = 678
Score = 28.5 bits (62), Expect = 5.9
Identities = 11/27 (40%), Positives = 20/27 (73%)
Frame = -3
Query: 286 LEVHISRKIGILIYLLVVLNLIFIFSF 312
L++ I+RK+ +L+Y LV L L+++ F
Sbjct: 118 LQIEINRKVVLLVYCLVTLYLLYLCFF 38
>TC211867 weakly similar to UP|PNX1_HUMAN (P55347) Homeobox protein PKNOX1
(PBX/knotted homeobox 1) (Homeobox protein PREP-1),
partial (6%)
Length = 900
Score = 28.5 bits (62), Expect = 5.9
Identities = 13/40 (32%), Positives = 20/40 (49%)
Frame = +3
Query: 34 NTARMHKKNCRQFAQHLKLIGNLLDQLKISELKKYPETRE 73
NT +H+K F KLI L + + ++ K+P T E
Sbjct: 459 NTLLLHEKQANNFQSERKLITALSETTEEPQIAKFPATEE 578
>TC214167 similar to UP|Q6NLW5 (Q6NLW5) At5g48720, partial (30%)
Length = 1252
Score = 28.5 bits (62), Expect = 5.9
Identities = 13/30 (43%), Positives = 18/30 (59%)
Frame = +1
Query: 271 HHKKKPKFQKRIRCQLEVHISRKIGILIYL 300
HH K F KRI + V +S K+G L+Y+
Sbjct: 514 HHVNKMWFNKRISLEPPVILSSKVGNLLYI 603
>TC216024 similar to GB|AAM78036.1|21928015|AY125526 At2g01100/F23H14.7
{Arabidopsis thaliana;} , partial (14%)
Length = 1433
Score = 28.5 bits (62), Expect = 5.9
Identities = 12/30 (40%), Positives = 17/30 (56%)
Frame = +2
Query: 252 SNNSHSDANGSKGHSSNEKHHKKKPKFQKR 281
S+ S DA KGH ++KHHK+ + R
Sbjct: 782 SDLSSDDAPRKKGHRKHQKHHKRSHNVELR 871
>TC216023 similar to GB|AAM78036.1|21928015|AY125526 At2g01100/F23H14.7
{Arabidopsis thaliana;} , partial (15%)
Length = 1437
Score = 28.5 bits (62), Expect = 5.9
Identities = 12/31 (38%), Positives = 17/31 (54%)
Frame = +3
Query: 252 SNNSHSDANGSKGHSSNEKHHKKKPKFQKRI 282
S+ S DA KGH + KHHK+ + R+
Sbjct: 780 SDLSSDDAPHKKGHRKHHKHHKRSHNVELRL 872
>TC207970 weakly similar to UP|Q94A73 (Q94A73) AT5g66560/K1F13_23, partial
(16%)
Length = 1114
Score = 28.5 bits (62), Expect = 5.9
Identities = 25/105 (23%), Positives = 47/105 (43%), Gaps = 15/105 (14%)
Frame = +1
Query: 118 IDRYLRLVPLITLVDNARATRVRERLEVIEKDQCAYTLDDEDQKVQTVF----------- 166
ID +L+ P ++ +D + V + + + ++ CA+ ++ VQTV
Sbjct: 304 IDIFLKAHPALSDMDRKKVCSVMD-CQKLSREACAHAAQNDRLPVQTVVQVLYYEQQRLR 480
Query: 167 --MKPEHDKEDTVVLKKTLSCS--YPNFSFTEALKKENEKLQLEL 207
M E +V K + + +P + L++ENE L+LEL
Sbjct: 481 NAMNGSRSGESSVDSKLNVYSTDLHPVSNELSTLRRENEDLKLEL 615
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.139 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,159,182
Number of Sequences: 63676
Number of extensions: 277387
Number of successful extensions: 1993
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 1974
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1988
length of query: 417
length of database: 12,639,632
effective HSP length: 100
effective length of query: 317
effective length of database: 6,272,032
effective search space: 1988234144
effective search space used: 1988234144
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC136504.6


