

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136472.17 - phase: 0
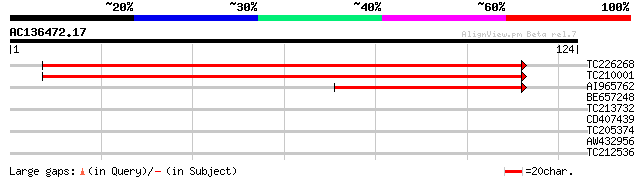
(124 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226268 similar to UP|Q9SXL2 (Q9SXL2) Serine decarboxylase, par... 200 1e-52
TC210001 similar to UP|Q9MA74 (Q9MA74) Histidine decarboxylase (... 196 3e-51
AI965762 50 3e-07
BE657248 26 4.1
TC213732 weakly similar to UP|Q6ID69 (Q6ID69) At3g09032, partial... 26 4.1
CD407439 25 7.0
TC205374 similar to UP|V714_ARATH (Q9FMR5) Vesicle-associated me... 25 7.0
AW432956 25 9.2
TC212536 homologue to UP|P93484 (P93484) BP-80 vacuolar sorting ... 25 9.2
>TC226268 similar to UP|Q9SXL2 (Q9SXL2) Serine decarboxylase, partial (81%)
Length = 1703
Score = 200 bits (509), Expect = 1e-52
Identities = 93/106 (87%), Positives = 101/106 (94%)
Frame = +1
Query: 8 FEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 67
F+KEVQKCLRNAHYFK RL+EAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV
Sbjct: 1207 FQKEVQKCLRNAHYFKGRLVEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 1386
Query: 68 VVMPNVTIEKLDDFLNELVQKRATWFEYGTFQPYCIASDVGENSCL 113
VVMPN+TIEKLDDFLNEL++KRATWF+ G QPYCI+SDVGE +CL
Sbjct: 1387 VVMPNITIEKLDDFLNELLEKRATWFQDGKDQPYCISSDVGEKNCL 1524
>TC210001 similar to UP|Q9MA74 (Q9MA74) Histidine decarboxylase (Serine
decarboxylase), partial (42%)
Length = 791
Score = 196 bits (497), Expect = 3e-51
Identities = 90/106 (84%), Positives = 98/106 (91%)
Frame = +2
Query: 8 FEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 67
F+KEVQKCLRNAHYFKDRL++AGIGAMLNELSSTVVFERPHDE F+ KWQLAC+GN+AHV
Sbjct: 341 FQKEVQKCLRNAHYFKDRLVDAGIGAMLNELSSTVVFERPHDEGFVHKWQLACQGNVAHV 520
Query: 68 VVMPNVTIEKLDDFLNELVQKRATWFEYGTFQPYCIASDVGENSCL 113
VVMPNVTIEKLDDFLNELVQKRA WF G QPYCIASDVG+ +CL
Sbjct: 521 VVMPNVTIEKLDDFLNELVQKRAVWFRDGNCQPYCIASDVGQENCL 658
>AI965762
Length = 311
Score = 50.1 bits (118), Expect = 3e-07
Identities = 24/42 (57%), Positives = 30/42 (71%)
Frame = +1
Query: 72 NVTIEKLDDFLNELVQKRATWFEYGTFQPYCIASDVGENSCL 113
N+TI KLDD LNEL++K AT E G PYCI+ D G+ +CL
Sbjct: 10 NITIAKLDDSLNELLEKPATCVEDGEDHPYCISLDGGD*TCL 135
>BE657248
Length = 746
Score = 26.2 bits (56), Expect = 4.1
Identities = 13/33 (39%), Positives = 18/33 (54%)
Frame = +2
Query: 90 ATWFEYGTFQPYCIASDVGENSCLAYVLIISNW 122
+TWF Y F C+ G N YVL+++NW
Sbjct: 488 STWFGY-QFSTSCL----GRNFNTRYVLLLANW 571
>TC213732 weakly similar to UP|Q6ID69 (Q6ID69) At3g09032, partial (18%)
Length = 550
Score = 26.2 bits (56), Expect = 4.1
Identities = 13/38 (34%), Positives = 18/38 (47%)
Frame = -2
Query: 73 VTIEKLDDFLNELVQKRATWFEYGTFQPYCIASDVGEN 110
+T E + FL +L+ T TF P C + GEN
Sbjct: 231 LTKEDEETFLEDLLLMETTLIALATFAPSCCKNLTGEN 118
>CD407439
Length = 413
Score = 25.4 bits (54), Expect = 7.0
Identities = 13/48 (27%), Positives = 23/48 (47%)
Frame = +1
Query: 38 LSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPNVTIEKLDDFLNEL 85
LSS +FE+ IR WQL + +++ N+ + F+N +
Sbjct: 268 LSSAYIFEKDLHSSSIRAWQLNYWAT*SPLILTSNIKYQISGVFMNSV 411
>TC205374 similar to UP|V714_ARATH (Q9FMR5) Vesicle-associated membrane
protein 714 (AtVAMP714), partial (98%)
Length = 1159
Score = 25.4 bits (54), Expect = 7.0
Identities = 14/43 (32%), Positives = 24/43 (55%), Gaps = 1/43 (2%)
Frame = -2
Query: 39 SSTVVFERPHDEEF-IRKWQLACKGNIAHVVVMPNVTIEKLDD 80
+STVVF +PH + F +R+ +G + H V + E ++D
Sbjct: 390 NSTVVFHKPHLDIFQVRERNSPSEGVVGHTDVGDAIRSEYVED 262
>AW432956
Length = 430
Score = 25.0 bits (53), Expect = 9.2
Identities = 8/18 (44%), Positives = 12/18 (66%)
Frame = +2
Query: 4 LPYSFEKEVQKCLRNAHY 21
+P + K +KCL+N HY
Sbjct: 185 VPVALHKAYEKCLQNVHY 238
>TC212536 homologue to UP|P93484 (P93484) BP-80 vacuolar sorting receptor,
partial (30%)
Length = 567
Score = 25.0 bits (53), Expect = 9.2
Identities = 16/59 (27%), Positives = 28/59 (47%), Gaps = 6/59 (10%)
Frame = +3
Query: 61 KGNIAHVVVMPNVTIE------KLDDFLNELVQKRATWFEYGTFQPYCIASDVGENSCL 113
KG+ V ++P + + KL+ +++ + FE T C++SDV N CL
Sbjct: 294 KGSRGDVTILPTLVVNNRQYRGKLEK--GAVLKAICSGFEETTEPTVCLSSDVETNECL 464
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.138 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,253,108
Number of Sequences: 63676
Number of extensions: 73846
Number of successful extensions: 413
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 411
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 413
length of query: 124
length of database: 12,639,632
effective HSP length: 85
effective length of query: 39
effective length of database: 7,227,172
effective search space: 281859708
effective search space used: 281859708
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 53 (25.0 bits)
Medicago: description of AC136472.17


