

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136472.15 + phase: 0
(486 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
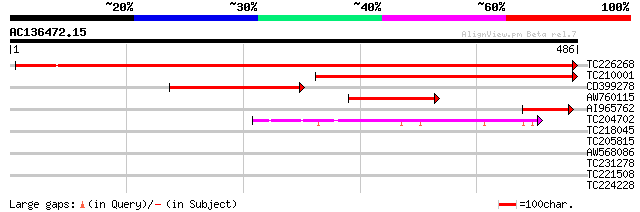
Score E
Sequences producing significant alignments: (bits) Value
TC226268 similar to UP|Q9SXL2 (Q9SXL2) Serine decarboxylase, par... 889 0.0
TC210001 similar to UP|Q9MA74 (Q9MA74) Histidine decarboxylase (... 427 e-120
CD399278 202 3e-52
AW760115 105 6e-23
AI965762 59 6e-09
TC204702 similar to UP|Q6Q4I2 (Q6Q4I2) Glutamate decarboxylase 4... 52 5e-07
TC218045 similar to UP|DCE_LYCES (P54767) Glutamate decarboxylas... 32 0.83
TC205815 similar to UP|Q9SUY5 (Q9SUY5) Dihydrolipoamide S-acetyl... 31 1.4
AW568086 30 3.2
TC231278 29 4.1
TC221508 similar to UP|Q8LF92 (Q8LF92) Homoserine dehydrogenase-... 28 9.2
TC224228 28 9.2
>TC226268 similar to UP|Q9SXL2 (Q9SXL2) Serine decarboxylase, partial (81%)
Length = 1703
Score = 889 bits (2297), Expect = 0.0
Identities = 428/481 (88%), Positives = 455/481 (93%)
Frame = +1
Query: 6 DVLVNGLSTNGAVELLPDDFDVSAIIKDPVPPVVAADNGIGKEEAKINGGKEKREIVLGR 65
D L L NGAVE LP+DFD +A+I DPVP V DNGI KEEA+I GKEKREIVLGR
Sbjct: 100 DALNEDLRINGAVEPLPEDFDATAVIIDPVPLAVV-DNGIVKEEAQIIKGKEKREIVLGR 276
Query: 66 NIHTTCLEVTEPEADDEITGDRDAHMASVLARYRKSLTERTKYHLGYPYNLDFDYGALSQ 125
NIHT+CLEVTEPEADDE+TGDR+AHMASVLARY+++LTERTK+HLGYPYNLDFDYGAL+Q
Sbjct: 277 NIHTSCLEVTEPEADDEVTGDREAHMASVLARYKRALTERTKHHLGYPYNLDFDYGALTQ 456
Query: 126 LQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEGNLHGI 185
LQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEGNLHGI
Sbjct: 457 LQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEGNLHGI 636
Query: 186 LVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKP 245
LVGREV PDGILYASRESHYS+FKAARMYRMECEKV+TL SGEIDCDDFKAKLL HQDKP
Sbjct: 637 LVGREVFPDGILYASRESHYSVFKAARMYRMECEKVDTLWSGEIDCDDFKAKLLSHQDKP 816
Query: 246 AIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFK 305
AIINVNIGTTVKGAVDDLDLVI+KLEEAGFS DRFYIH DGALFGLMMPFVKRAPKVTFK
Sbjct: 817 AIINVNIGTTVKGAVDDLDLVIKKLEEAGFSHDRFYIHCDGALFGLMMPFVKRAPKVTFK 996
Query: 306 KPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLASRDATIMGSRNGHAPIFLW 365
KP+GSVSVSGHKFVGCPMPCGVQITRLE++NAL+R+VEYLASRDATIMGSRNGHAPIFLW
Sbjct: 997 KPVGSVSVSGHKFVGCPMPCGVQITRLEYVNALARDVEYLASRDATIMGSRNGHAPIFLW 1176
Query: 366 YTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQ 425
YTLNRKGYRGFQKEVQKCLRNAHYFK RL+EAGIGAMLNELSSTVVFERPHDEEFIRKWQ
Sbjct: 1177YTLNRKGYRGFQKEVQKCLRNAHYFKGRLVEAGIGAMLNELSSTVVFERPHDEEFIRKWQ 1356
Query: 426 LACKGNIAHVVVMPNVTIEKLDDFLNELVQKRATWFEDGTFQPYCIASDVGENSCLCAQH 485
LACKGNIAHVVVMPN+TIEKLDDFLNEL++KRATWF+DG QPYCI+SDVGE +CLCA H
Sbjct: 1357LACKGNIAHVVVMPNITIEKLDDFLNELLEKRATWFQDGKDQPYCISSDVGEKNCLCALH 1536
Query: 486 K 486
K
Sbjct: 1537K 1539
>TC210001 similar to UP|Q9MA74 (Q9MA74) Histidine decarboxylase (Serine
decarboxylase), partial (42%)
Length = 791
Score = 427 bits (1099), Expect = e-120
Identities = 200/224 (89%), Positives = 214/224 (95%)
Frame = +2
Query: 263 LDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCP 322
LDLVI+KLEEAGFSQDRFYIH DGALFGLM+PFVKRAPK++FKKPIGSVSVSGHKFVGCP
Sbjct: 2 LDLVIKKLEEAGFSQDRFYIHCDGALFGLMLPFVKRAPKISFKKPIGSVSVSGHKFVGCP 181
Query: 323 MPCGVQITRLEHINALSRNVEYLASRDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQK 382
MPCGVQITRLEH+NALSRNVEYLASRDATIMGSRNGHAPIFLWY+LN KGYRGFQKEVQK
Sbjct: 182 MPCGVQITRLEHVNALSRNVEYLASRDATIMGSRNGHAPIFLWYSLNMKGYRGFQKEVQK 361
Query: 383 CLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPNVT 442
CLRNAHYFKDRL++AGIGAMLNELSSTVVFERPHDE F+ KWQLAC+GN+AHVVVMPNVT
Sbjct: 362 CLRNAHYFKDRLVDAGIGAMLNELSSTVVFERPHDEGFVHKWQLACQGNVAHVVVMPNVT 541
Query: 443 IEKLDDFLNELVQKRATWFEDGTFQPYCIASDVGENSCLCAQHK 486
IEKLDDFLNELVQKRA WF DG QPYCIASDVG+ +CLCA H+
Sbjct: 542 IEKLDDFLNELVQKRAVWFRDGNCQPYCIASDVGQENCLCALHR 673
>CD399278
Length = 595
Score = 202 bits (514), Expect = 3e-52
Identities = 95/115 (82%), Positives = 104/115 (89%)
Frame = -1
Query: 138 FIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEGNLHGILVGREVLPDGIL 197
FIESNYGVHS QFEVGVLDWFA LWELEK+EYWGYITN GTEGNLHGILVGREV P+GIL
Sbjct: 595 FIESNYGVHSSQFEVGVLDWFAWLWELEKDEYWGYITNYGTEGNLHGILVGREVFPNGIL 416
Query: 198 YASRESHYSIFKAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNI 252
YAS+ESHYS+FKAARMY MEC K+ TL SGEIDCD F+AKLL H+DKP I++VNI
Sbjct: 415 YASQESHYSVFKAARMYIMECVKINTLWSGEIDCDGFEAKLLCHKDKPGIVDVNI 251
>AW760115
Length = 235
Score = 105 bits (261), Expect = 6e-23
Identities = 53/78 (67%), Positives = 58/78 (73%)
Frame = +2
Query: 291 LMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLASRDA 350
LMMPF KRAP+VT KKP SVS SGHKFVGCPMPCGV ITRL ++NAL+ +VEYLA RDA
Sbjct: 2 LMMPFRKRAPEVTIKKPGVSVSASGHKFVGCPMPCGVLITRL*YVNALAIDVEYLACRDA 181
Query: 351 TIMGSRNGHAPIFLWYTL 368
T M FLWY L
Sbjct: 182 TFMDWSEWSCAHFLWYAL 235
>AI965762
Length = 311
Score = 58.5 bits (140), Expect = 6e-09
Identities = 27/44 (61%), Positives = 33/44 (74%)
Frame = +1
Query: 440 NVTIEKLDDFLNELVQKRATWFEDGTFQPYCIASDVGENSCLCA 483
N+TI KLDD LNEL++K AT EDG PYCI+ D G+ +CLCA
Sbjct: 10 NITIAKLDDSLNELLEKPATCVEDGEDHPYCISLDGGD*TCLCA 141
>TC204702 similar to UP|Q6Q4I2 (Q6Q4I2) Glutamate decarboxylase 4a , partial
(91%)
Length = 1814
Score = 52.4 bits (124), Expect = 5e-07
Identities = 67/269 (24%), Positives = 124/269 (45%), Gaps = 21/269 (7%)
Frame = +3
Query: 209 KAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDL----D 264
K AR + +E ++V+ L D KA L ++ + + +G+T+ G +D+ D
Sbjct: 573 KFARYFEVELKEVK-LRDDYYVMDPEKAVELVDENTICVAAI-LGSTLNGEFEDVKRLND 746
Query: 265 LVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFK-KPIGSVSVSGHKFVGCPM 323
L+I+K + G+ IHVD A G + PF+ + F+ + + S++VSGHK+
Sbjct: 747 LLIEKNKITGWDTP---IHVDAASGGFIAPFIYPELEWDFRLQLVKSINVSGHKYGLVYA 917
Query: 324 PCGVQITRLEH--INALSRNVEYLASRDA--TIMGSRNGHAPIFLWYTLNRKGYRGFQKE 379
G I R + L ++ YL + T+ S+ I +Y L R G+ G++
Sbjct: 918 GIGWVIWRSKQDLPEELIFHINYLGADQPTFTLNFSKGSSQVIAQYYQLIRLGFEGYRNV 1097
Query: 380 VQKCLRNAHYFKDRLIEAGIGAMLNE-----LSSTVVFERPHDEEFIRKWQLACKGNIAH 434
++ C N K+ L + G +++++ L + + + H +EF L G I
Sbjct: 1098MENCRDNMLVLKEGLEKTGRFSIVSKDNGVPLVAFTLKDHTHFDEFQISDFLRRFGWIVP 1277
Query: 435 VVVMP----NVTIEKL---DDFLNELVQK 456
MP +VT+ ++ +DF L ++
Sbjct: 1278AYTMPPDAQHVTVLRVVIREDFSRTLAER 1364
>TC218045 similar to UP|DCE_LYCES (P54767) Glutamate decarboxylase (GAD)
(ERT D1) , partial (49%)
Length = 740
Score = 31.6 bits (70), Expect = 0.83
Identities = 26/92 (28%), Positives = 47/92 (50%), Gaps = 4/92 (4%)
Frame = +1
Query: 209 KAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDL----D 264
K AR + +E ++V+ L G D KA + ++ + + +G+T+ G +D+ +
Sbjct: 463 KFARYFEVELKEVK-LKEGYYVMDPAKAVEMVDENTICVAAI-LGSTMTGEFEDVKLLNE 636
Query: 265 LVIQKLEEAGFSQDRFYIHVDGALFGLMMPFV 296
L+ +K E G+ IHVD A G + PF+
Sbjct: 637 LLTKKNNETGWDTP---IHVDAASGGFIAPFL 723
>TC205815 similar to UP|Q9SUY5 (Q9SUY5) Dihydrolipoamide S-acetyltransferase,
partial (60%)
Length = 1893
Score = 30.8 bits (68), Expect = 1.4
Identities = 32/115 (27%), Positives = 48/115 (40%)
Frame = +2
Query: 21 LPDDFDVSAIIKDPVPPVVAADNGIGKEEAKINGGKEKREIVLGRNIHTTCLEVTEPEAD 80
L + +DV A + D + VVAA EA EK E++L +I + TE
Sbjct: 776 LKEQYDVKASVNDIIIKVVAAALR-NVPEANAYWNVEKDEVILNDSIDISIAVATEKGLM 952
Query: 81 DEITGDRDAHMASVLARYRKSLTERTKYHLGYPYNLDFDYGALSQLQHFSINNLG 135
I + D S ++ K L + + P+ +F G FSI+NLG
Sbjct: 953 TPIIKNADQKTISAISSEVKELAAKARAGKLKPH--EFQGGT------FSISNLG 1093
>AW568086
Length = 435
Score = 29.6 bits (65), Expect = 3.2
Identities = 16/41 (39%), Positives = 21/41 (51%)
Frame = +3
Query: 369 NRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELSST 409
NR GY F E L+ H+ KDR I IG + N+L +
Sbjct: 51 NRSGYNFFFAEQHARLKLLHHGKDREISRMIGELWNKLKES 173
>TC231278
Length = 614
Score = 29.3 bits (64), Expect = 4.1
Identities = 18/55 (32%), Positives = 23/55 (41%), Gaps = 9/55 (16%)
Frame = -2
Query: 159 ARLWELEKNEYWGYITNC---------GTEGNLHGILVGREVLPDGILYASRESH 204
A+ W + N YW Y T+C G L+ I GRE D YA + H
Sbjct: 460 AKSWTVT*NHYW*YRTSCFIISY*YKIGKTSALYSIEYGREKKTDKFAYAD*Q*H 296
>TC221508 similar to UP|Q8LF92 (Q8LF92) Homoserine dehydrogenase-like
protein, partial (60%)
Length = 912
Score = 28.1 bits (61), Expect = 9.2
Identities = 24/86 (27%), Positives = 39/86 (44%), Gaps = 1/86 (1%)
Frame = +2
Query: 33 DPVPPVVAADNG-IGKEEAKINGGKEKREIVLGRNIHTTCLEVTEPEADDEITGDRDAHM 91
DPV ++ + +G +G +++ GK ++V L TEP+ D++ G A
Sbjct: 116 DPVHQIIGSLSGTLGYVMSEVEDGKPLSQVVRDAK----SLGYTEPDPRDDLGGMDVARK 283
Query: 92 ASVLARYRKSLTERTKYHLGYPYNLD 117
A +LAR LG+ NLD
Sbjct: 284 ALILARI-----------LGHQINLD 328
>TC224228
Length = 437
Score = 28.1 bits (61), Expect = 9.2
Identities = 24/95 (25%), Positives = 39/95 (40%)
Frame = +1
Query: 38 VVAADNGIGKEEAKINGGKEKREIVLGRNIHTTCLEVTEPEADDEITGDRDAHMASVLAR 97
V++ + +G EE GK L +I+ L VT DA+ VL
Sbjct: 55 VISQWSKLGLEEVDFGEGKPLHMGPLTSDIYCLFLPVTG-----------DANAVRVLVS 201
Query: 98 YRKSLTERTKYHLGYPYNLDFDYGALSQLQHFSIN 132
+S+ ER +YH+ + + G +QH + N
Sbjct: 202 VPESMVERFQYHMNESWQGKEENGNHHDVQHHNEN 306
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.139 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,956,719
Number of Sequences: 63676
Number of extensions: 274729
Number of successful extensions: 1181
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1177
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1180
length of query: 486
length of database: 12,639,632
effective HSP length: 101
effective length of query: 385
effective length of database: 6,208,356
effective search space: 2390217060
effective search space used: 2390217060
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC136472.15


