

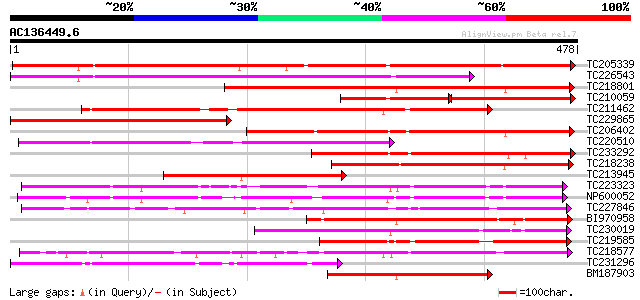
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136449.6 - phase: 0
(478 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC205339 weakly similar to UP|Q6QDB6 (Q6QDB6) UDP-glucose glucos... 521 e-148
TC226543 weakly similar to UP|Q6VAA9 (Q6VAA9) UDP-glycosyltransf... 313 1e-85
TC218801 similar to UP|Q8W3P8 (Q8W3P8) ABA-glucosyltransferase, ... 309 2e-84
TC210059 similar to UP|Q8S9A1 (Q8S9A1) Glucosyltransferase-8 (Fr... 172 2e-84
TC211462 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltran... 304 5e-83
TC229865 weakly similar to UP|P93789 (P93789) UDP-glucose glucos... 303 1e-82
TC206402 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, p... 265 3e-71
TC220510 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltran... 244 6e-65
TC233292 similar to UP|Q8S9A0 (Q8S9A0) Glucosyltransferase-9, pa... 234 8e-62
TC218238 weakly similar to UP|Q8H0F2 (Q8H0F2) Anthocyanin 3'-glu... 212 3e-55
TC213945 similar to UP|Q6QDB6 (Q6QDB6) UDP-glucose glucosyltrans... 209 2e-54
TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransfer... 204 9e-53
NP600052 zeatin O-glucosyltransferase [Glycine max] 201 8e-52
TC227846 weakly similar to UP|Q947K4 (Q947K4) Thiohydroximate S-... 193 2e-49
BI970958 178 4e-45
TC230019 weakly similar to UP|Q6WFW1 (Q6WFW1) Glucosyltransferas... 178 4e-45
TC219585 weakly similar to UP|Q8L5C7 (Q8L5C7) UDP-glucuronosyltr... 174 6e-44
TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (F... 174 6e-44
TC231296 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltran... 173 1e-43
BM187903 172 3e-43
>TC205339 weakly similar to UP|Q6QDB6 (Q6QDB6) UDP-glucose
glucosyltransferase, partial (55%)
Length = 1537
Score = 521 bits (1343), Expect = e-148
Identities = 261/486 (53%), Positives = 349/486 (71%), Gaps = 11/486 (2%)
Frame = +2
Query: 3 NENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGK---AKI 59
N + ELHI+ FPF GH+IP D+AR F+ RG++ TIVTT LNV I TIGK I
Sbjct: 14 NMDGELHIMLFPFPGQGHLIPMSDMARAFNGRGVRTTIVTTPLNVATIRGTIGKETETDI 193
Query: 60 NIKTIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDCLVAD 119
I T+KFPS E GLPEGCEN+ES +PD + F+K+ +L PLEH+L Q +P CL+A
Sbjct: 194 EILTVKFPSAE-AGLPEGCENTESIPSPDLVLTFLKAIRMLEAPLEHLLLQHRPHCLIAS 370
Query: 120 MFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEIT 179
FFPW++ SA K IPR+VFHG G F LC C R Y+P VSS T+PF++P+LPG+I
Sbjct: 371 AFFPWASHSATKLKIPRLVFHGTGVFALCASECVRLYQPHKNVSSDTDPFIIPHLPGDIQ 550
Query: 180 LTKMQLPQLPQHD----KVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNEL--- 232
+T++ LP + D T++L+E ESE+ S+G+I N+FYELE VYAD+Y +L
Sbjct: 551 MTRLLLPDYAKTDGDGETGLTRVLQEIKESELASYGMIVNSFYELEQVYADYYDKQLLQV 730
Query: 233 -GRKAWHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSD 291
GR+AW++GP+SLCN+D K RG++AS+D+ + LKWL SK+ NSV+YVCFGS+ FS+
Sbjct: 731 QGRRAWYIGPLSLCNQD---KGKRGKQASVDQGDILKWLDSKKANSVVYVCFGSIANFSE 901
Query: 292 AQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVM 351
QL+EIA GLE S FIWVVR+S K ++ WLPEGFE R G+G+II GWAPQV+
Sbjct: 902 TQLREIARGLEDSGQQFIWVVRRSDK---DDKGWLPEGFETRTTSEGRGVIIWGWAPQVL 1072
Query: 352 ILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTW 411
ILDH++VG FVTHCGWNSTLE VSAG+PM+TWP+ EQFYN KF++DI++IGV VGV+ W
Sbjct: 1073ILDHQAVGAFVTHCGWNSTLEAVSAGVPMLTWPVSAEQFYNEKFVTDILQIGVPVGVKKW 1252
Query: 412 IGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIE 471
+ G+ + + ++KA+ RIM+G+EAE MR+RA + +MA A++ GSSY F++LI+
Sbjct: 1253NRI-VGDNITSNALQKALHRIMIGEEAEPMRNRAHKLAQMATTALQHNGSSYCHFTHLIQ 1429
Query: 472 DLKSRA 477
L+S A
Sbjct: 1430HLRSIA 1447
>TC226543 weakly similar to UP|Q6VAA9 (Q6VAA9) UDP-glycosyltransferase 73E1,
partial (21%)
Length = 1237
Score = 313 bits (801), Expect = 1e-85
Identities = 165/399 (41%), Positives = 238/399 (59%), Gaps = 7/399 (1%)
Frame = +2
Query: 1 MTNENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGK---- 56
M + EL IF PFL+ HIIP VD+AR+F+ + VTI+TT N + ++I
Sbjct: 26 MEKKKGELKSIFLPFLSTSHIIPLVDMARLFALHDVDVTIITTAHNATVFQKSIDLDASR 205
Query: 57 -AKINIKTIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDC 115
I + FP+ + GLP G E + + LL++ E + +PD
Sbjct: 206 GRPIRTHVVNFPAAQ-VGLPVGIEAFNVDTPREMTPRIYMGLSLLQQVFEKLFHDLQPDF 382
Query: 116 LVADMFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLP 175
+V DMF PWS D+AAK IPRI+FHG + QY P + T+ FV+P LP
Sbjct: 383 IVTDMFHPWSVDAAAKLGIPRIMFHGASYLARSAAHSVEQYAPHLEAKFDTDKFVLPGLP 562
Query: 176 GEITLTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRK 235
+ +T++QLP + +T+L+ +SE KS+G + N+FY+LE Y +HY++ +G K
Sbjct: 563 DNLEMTRLQLPDWLRSPNQYTELMRTIKQSEKKSYGSLFNSFYDLESAYYEHYKSIMGTK 742
Query: 236 AWHLGPVSL-CNRDTEEKACRGREASIDEHE-CLKWLQSKEPNSVIYVCFGSMTVFSDAQ 293
+W +GPVSL N+D ++KA RG +E E LKWL SK +SV+YV FGS+ F +Q
Sbjct: 743 SWGIGPVSLWANQDAQDKAARGYAKEEEEKEGWLKWLNSKAESSVLYVSFGSINKFPYSQ 922
Query: 294 LKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMIL 353
L EIA LE S FIWVVRK+ EG+N E FE+R++ S KG +I GWAPQ++IL
Sbjct: 923 LVEIARALEDSGHDFIWVVRKNDGGEGDN---FLEEFEKRMKESNKGYLIWGWAPQLLIL 1093
Query: 354 DHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYN 392
++ ++GG V+HCGWN+ +E V++ LPM TWP++ E F+N
Sbjct: 1094ENPAIGGLVSHCGWNTVVESVNSVLPMATWPLFAEHFFN 1210
>TC218801 similar to UP|Q8W3P8 (Q8W3P8) ABA-glucosyltransferase, partial
(60%)
Length = 1281
Score = 309 bits (792), Expect = 2e-84
Identities = 153/303 (50%), Positives = 211/303 (69%), Gaps = 8/303 (2%)
Frame = +1
Query: 182 KMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWHLGP 241
+M QLP + ++ + + E KSFG N+F++LEP YA+ +N+ G+KAW +GP
Sbjct: 1 EMTRSQLPVFLRTPSQFPDRVRQLEEKSFGTFVNSFHDLEPAYAEQVKNKWGKKAWIIGP 180
Query: 242 VSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGL 301
VSLCNR E+K RG+ +IDE +CL WL SK+PNSV+YV FGS+ QLKEIA GL
Sbjct: 181 VSLCNRTAEDKTERGKLPTIDEEKCLNWLNSKKPNSVLYVSFGSLLRLPSEQLKEIACGL 360
Query: 302 EASEVPFIWVVRKSAKSEGENLE-----WLPEGFEERIEGSGKGLIIRGWAPQVMILDHE 356
EASE FIWVVR + EN E +LPEGFE+R++ KGL++RGWAPQ++IL+H
Sbjct: 361 EASEQSFIWVVRNIHNNPSENKENGNGNFLPEGFEQRMKEKDKGLVLRGWAPQLLILEHV 540
Query: 357 SVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGG 416
++ GF+THCGWNSTLE V AG+PM+TWP+ EQF N K +++++KIGV VG + W+
Sbjct: 541 AIKGFMTHCGWNSTLESVCAGVPMITWPLSAEQFSNEKLITEVLKIGVQVGSREWLSWNS 720
Query: 417 --GEPVKKDVIEKAVRRIMV-GDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDL 473
+ V ++ +E AVR++MV +EAEEM +R K+ + A+RAVE GG+SY D LIE+L
Sbjct: 721 EWKDLVGREKVESAVRKLMVESEEAEEMTTRVKDIAEKAKRAVEEGGTSYADAEALIEEL 900
Query: 474 KSR 476
K+R
Sbjct: 901 KAR 909
>TC210059 similar to UP|Q8S9A1 (Q8S9A1) Glucosyltransferase-8 (Fragment),
partial (37%)
Length = 803
Score = 172 bits (436), Expect(2) = 2e-84
Identities = 83/105 (79%), Positives = 95/105 (90%)
Frame = +2
Query: 373 GVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGEPVKKDVIEKAVRRI 432
G AG+PMVTWPMY EQFYNAKFL+DIVKIGV VGVQTWIGM G +PVKK+ +EKAVRRI
Sbjct: 272 GCVAGVPMVTWPMYSEQFYNAKFLTDIVKIGVSVGVQTWIGMMGRDPVKKEPVEKAVRRI 451
Query: 433 MVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDLKSRA 477
MVG+EAEEMR+RAKE +MA+RAVE GGSSYNDF++LIEDL+SRA
Sbjct: 452 MVGEEAEEMRNRAKELARMAKRAVEEGGSSYNDFNSLIEDLRSRA 586
Score = 159 bits (401), Expect(2) = 2e-84
Identities = 77/95 (81%), Positives = 80/95 (84%)
Frame = +1
Query: 280 YVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGK 339
Y+CFG MT FSDAQLKEIA+GLEAS FIWVV+K E LEWLPEGFEERI G GK
Sbjct: 1 YLCFGRMTAFSDAQLKEIALGLEASGQNFIWVVKKGLN---EKLEWLPEGFEERILGQGK 171
Query: 340 GLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEGV 374
GLIIRGWAPQVMILDHESVGGFVTHCGWNS LEGV
Sbjct: 172 GLIIRGWAPQVMILDHESVGGFVTHCGWNSVLEGV 276
>TC211462 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (64%)
Length = 1066
Score = 304 bits (779), Expect = 5e-83
Identities = 161/353 (45%), Positives = 216/353 (60%), Gaps = 6/353 (1%)
Frame = +3
Query: 61 IKTIKFPSPEETGLPEGCENSESALAPDKFIK-FMKSTLLLREPLEHVLEQEKPDCLVAD 119
+ T FPS EE GLP+G EN + +K + ++ +T LLREP+E +E++ PDC+VAD
Sbjct: 33 VHTFDFPS-EEVGLPDGVENLSAVTDLEKSYRIYIAATTLLREPIESFVERDPPDCIVAD 209
Query: 120 MFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEIT 179
+ W D A K IP + F+G F +C + ++++ D PFV+P+ P +T
Sbjct: 210 FLYCWVEDLAKKLRIPWLDFNGFSLFSICAMESVKKHRIGDG------PFVIPDFPDHVT 371
Query: 180 LTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELE-PVYADHYRNELGRKAWH 238
+ K + LE + +KS G I N F EL+ Y HY G KAWH
Sbjct: 372 IKSTP-------PKDMREFLEPLLTAALKSNGFIINNFAELDGEEYLRHYEKTTGHKAWH 530
Query: 239 LGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIA 298
LGP SL R EKA RG+++ + HECL WL SK NSV+YV FGS+ F D QL EIA
Sbjct: 531 LGPASLVRRTEMEKAERGQKSVVSTHECLSWLDSKRVNSVVYVSFGSLCYFPDKQLYEIA 710
Query: 299 MGLEASEVPFIWVVR----KSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMILD 354
G+EAS FIWVV K +SE E +WLP+GFEER KG+II+GWAPQV+IL+
Sbjct: 711 CGMEASGYEFIWVVPEKKGKEEESEEEKEKWLPKGFEER----KKGMIIKGWAPQVVILE 878
Query: 355 HESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVG 407
H +VG F+THCGWNST+E VSAG+PM+TWP++ +QFYN K ++ + IG G G
Sbjct: 879 HPAVGAFLTHCGWNSTVEAVSAGVPMITWPVHSDQFYNEKLITQVRGIGGGGG 1037
>TC229865 weakly similar to UP|P93789 (P93789) UDP-glucose
glucosyltransferase , partial (15%)
Length = 647
Score = 303 bits (776), Expect = 1e-82
Identities = 139/187 (74%), Positives = 166/187 (88%)
Frame = +1
Query: 1 MTNENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKIN 60
M NEN+ELH++FFPF ANGHIIP +DLARVF+SRG+K T+VTT LNVPLISRTIGKA I
Sbjct: 85 MGNENRELHVLFFPFPANGHIIPSIDLARVFASRGIKTTVVTTPLNVPLISRTIGKANIK 264
Query: 61 IKTIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDCLVADM 120
IKTIKFPS EETGLPEGCENS+SAL+ D + F+K+T+LLR+PLE++++QE PDC++ADM
Sbjct: 265 IKTIKFPSHEETGLPEGCENSDSALSSDLIMTFLKATVLLRDPLENLMQQEHPDCVIADM 444
Query: 121 FFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEITL 180
F+PW+TDSAAKF IPR+VFHG+GFFP CV AC R YKPQD VSS++EPF VP LPGEIT+
Sbjct: 445 FYPWATDSAAKFGIPRVVFHGMGFFPTCVSACVRTYKPQDNVSSWSEPFAVPELPGEITI 624
Query: 181 TKMQLPQ 187
TKMQLPQ
Sbjct: 625 TKMQLPQ 645
>TC206402 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, partial (59%)
Length = 1152
Score = 265 bits (677), Expect = 3e-71
Identities = 135/283 (47%), Positives = 193/283 (67%), Gaps = 6/283 (2%)
Frame = +2
Query: 200 EESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWHLGPVSLCNRDTEEKACRGREA 259
EE E+E+ S+GV+ N+F ELEP YA Y+ G K W +GPVSL N+D +KA RG A
Sbjct: 26 EEIREAEMSSYGVVMNSFEELEPAYATGYKKIRGDKLWCIGPVSLINKDHLDKAQRGT-A 202
Query: 260 SIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVRKSAKSE 319
SID + +KWL ++P +VIY C GS+ + QLKE+ + LEAS+ PFIWV+R+ SE
Sbjct: 203 SIDVSQHIKWLDCQKPGTVIYACLGSLCNLTTPQLKELGLALEASKRPFIWVIREGGHSE 382
Query: 320 GENLEWLPE-GFEERIEGSGKGLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGL 378
E +W+ E GFEER + + L+IRGWAPQ++IL H ++GGF+THCGWNSTLE + AG+
Sbjct: 383 -ELEKWIKEYGFEERT--NARSLLIRGWAPQILILSHPAIGGFITHCGWNSTLEAICAGV 553
Query: 379 PMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGG----GEPVKKDVIEKAVRRIM- 433
PM+TWP++ +QF N + ++K+GV VGV+ + G G VKK +E+A+ ++M
Sbjct: 554 PMLTWPLFADQFLNESLVVHVLKVGVKVGVEIPLTWGKEVEIGVQVKKKDVERAIAKLMD 733
Query: 434 VGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDLKSR 476
E+E+ R R +E +MA RAVE GGSSY++ + LI+D+ +
Sbjct: 734 ETSESEKRRKRVRELAEMANRAVEKGGSSYSNVTLLIQDIMQK 862
>TC220510 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (62%)
Length = 925
Score = 244 bits (623), Expect = 6e-65
Identities = 132/319 (41%), Positives = 188/319 (58%), Gaps = 2/319 (0%)
Frame = +3
Query: 8 LHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTI-GKAKINIKTIKF 66
L + F FLA GH+IP D+A +FS+RG VTI+TT N ++ +++ + + T++F
Sbjct: 9 LKLYFIHFLAEGHMIPLCDMATLFSTRGHHVTIITTPSNAQILRKSLPSHPLLRLHTVQF 188
Query: 67 PSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDCLVADMFFPWST 126
PS E GLP+G EN + D K +T +L+ P+E ++EQ+ PDC+VAD FPW
Sbjct: 189 PS-HEVGLPDGIENIYADSDLDSLRKVCSATAMLQPPIEDLVEQQPPDCIVADYLFPWVD 365
Query: 127 DSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEITLTKMQLP 186
D A K +IPR+ F+GL F +C + + S ++ ++ +LP ITL
Sbjct: 366 DLAKKLHIPRLAFNGLSLFTICAIHSS---------SESSDSTIIQSLPHPITLNATPPK 518
Query: 187 QLPQHDKVFTKLLEESNESEVKSFGVIANTFYELE-PVYADHYRNELGRKAWHLGPVSLC 245
+L TK LE E+E+KS+G+I N+F EL+ Y +Y G KAWHLGP SL
Sbjct: 519 EL-------TKFLETVLETELKSYGLIVNSFTELDGEEYTRYYEKTTGHKAWHLGPASLI 677
Query: 246 NRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEASE 305
R +EKA RG+++ + HEC+ WL SK NSV+Y+CFGS+ F D QL EIA G++AS
Sbjct: 678 GRTAQEKAERGQKSVVSMHECVAWLDSKRENSVVYICFGSLCYFQDKQLYEIACGIQASG 857
Query: 306 VPFIWVVRKSAKSEGENLE 324
FIWVV + E E E
Sbjct: 858 HDFIWVVPEKKGKEHEKEE 914
>TC233292 similar to UP|Q8S9A0 (Q8S9A0) Glucosyltransferase-9, partial (46%)
Length = 993
Score = 234 bits (596), Expect = 8e-62
Identities = 117/230 (50%), Positives = 167/230 (71%), Gaps = 7/230 (3%)
Frame = +1
Query: 255 RGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVRK 314
RG +ASI+EH CLKWL ++ SV+YVCFGS+ +QL E+A+ LE ++ PF+WV+R+
Sbjct: 214 RGDQASINEHHCLKWLDLQKSKSVVYVCFGSLCNLIPSQLVELALALEDTKRPFVWVIRE 393
Query: 315 SAKSEGENLEWLPE-GFEERIEGSGKGLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEG 373
+K + E +W+ E GFEER +G +GLIIRGWAPQV+IL H ++GGF+THCGWNSTLEG
Sbjct: 394 GSKYQ-ELEKWISEEGFEERTKG--RGLIIRGWAPQVLILSHHAIGGFLTHCGWNSTLEG 564
Query: 374 VSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGEP----VKKDVIEKAV 429
+ AGLPM+TWP++ +QF N K ++ ++KIGV VGV+ + G E VKK+ I +A+
Sbjct: 565 IGAGLPMITWPLFADQFLNEKLVTKVLKIGVSVGVEVPMKFGEEEKTGVLVKKEDINRAI 744
Query: 430 RRIM--VGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDLKSRA 477
+M G+E++E R RA + +MA+RAVE GGSS+ D S LI+D+ ++
Sbjct: 745 CMVMDDDGEESKERRERATKLSEMAKRAVENGGSSHLDLSLLIQDIMQQS 894
>TC218238 weakly similar to UP|Q8H0F2 (Q8H0F2) Anthocyanin
3'-glucosyltransferase, partial (26%)
Length = 936
Score = 212 bits (539), Expect = 3e-55
Identities = 106/207 (51%), Positives = 146/207 (70%), Gaps = 3/207 (1%)
Frame = +3
Query: 272 SKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVRKSAKSE-GENLEWLPEGF 330
SK NSV+YV FGSM F QL EIA LE S+ FIWVVRK +SE GE ++L E F
Sbjct: 6 SKTENSVLYVSFGSMNKFPTPQLVEIAHALEDSDHDFIWVVRKKGESEDGEGNDFLQE-F 182
Query: 331 EERIEGSGKGLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQF 390
++R++ S KG +I GWAPQ++IL+H ++G VTHCGWN+ +E V+AGLPM TWP++ EQF
Sbjct: 183 DKRVKASNKGYLIWGWAPQLLILEHHAIGAVVTHCGWNTIIESVNAGLPMATWPLFAEQF 362
Query: 391 YNAKFLSDIVKIGVGVGVQTWIGMG--GGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEF 448
YN K L+++++IGV VG + W G E VK++ I A+ +M G+E+ EMR RAK
Sbjct: 363 YNEKLLAEVLRIGVPVGAKEWRNWNEFGDEVVKREEIGNAIGVLMGGEESIEMRRRAKAL 542
Query: 449 GKMARRAVEVGGSSYNDFSNLIEDLKS 475
A++A++VGGSS+N+ LI++LKS
Sbjct: 543 SDAAKKAIQVGGSSHNNLKELIQELKS 623
>TC213945 similar to UP|Q6QDB6 (Q6QDB6) UDP-glucose glucosyltransferase,
partial (23%)
Length = 475
Score = 209 bits (532), Expect = 2e-54
Identities = 97/158 (61%), Positives = 122/158 (76%), Gaps = 3/158 (1%)
Frame = +2
Query: 130 AKFNIPRIVFHGLGFFPLCVLACTRQYKPQDK-VSSYTEPFVVPNLPGEITLTKMQLPQL 188
AKF IPR+VFHG FF LCV C Y+P DK SS ++ F++PN PGEI + K ++P
Sbjct: 2 AKFGIPRLVFHGTSFFSLCVTTCMPFYEPHDKYASSDSDSFLIPNFPGEIRIEKTKIPPY 181
Query: 189 PQHDKV--FTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWHLGPVSLCN 246
+ + KLLEE+ ESE++S+GV+ N+FYELE VYADH+RN LGRKAWH+GP+SLCN
Sbjct: 182 SKSKEKAGLAKLLEEAKESELRSYGVVVNSFYELEKVYADHFRNVLGRKAWHIGPLSLCN 361
Query: 247 RDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFG 284
+D EEKA RG+EASIDEHECLKWL +K+PNSVIY+C G
Sbjct: 362 KDAEEKARRGKEASIDEHECLKWLNTKKPNSVIYICXG 475
>TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransferase, partial
(89%)
Length = 1386
Score = 204 bits (518), Expect = 9e-53
Identities = 147/471 (31%), Positives = 226/471 (47%), Gaps = 11/471 (2%)
Frame = +1
Query: 11 IFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKINIKTIKFPSPE 70
+ PF A GH+ + L+R+ S + V V T ++ ++ + NI F P
Sbjct: 49 VLIPFPAQGHLNQLLHLSRLILSHNIPVHYVGTVTHIRQVTLRDHNSISNIHFHAFEVPS 228
Query: 71 ETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLE----QEKPDCLVADMFFPWST 126
P N E+ P + +++ LREP+ +L Q K ++ D
Sbjct: 229 FVSPPPNPNNEETDF-PAHLLPSFEASSHLREPVRKLLHSLSSQAKRVIVIHDSVMASVA 405
Query: 127 DSAAKF-NIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEITLTKMQL 185
A N+ FH F V + +P V P +P++ G T M
Sbjct: 406 QDATNMPNVENYTFHSTCTFGTAVFYWDKMGRPL--VDGMLVP-EIPSMEGCFTTDFMNF 576
Query: 186 PQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHY-RNELGRKAWHLGPVSL 244
+ Q D F K+ + G I NT +E Y + R G+K W LGP +
Sbjct: 577 -MIAQRD--FRKVND----------GNIYNTSRAIEGAYIEWMERFTGGKKLWALGPFNP 717
Query: 245 CNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEAS 304
+ ++ S + H CL+WL ++PNSV+YV FG+ T F + Q+K+IA GLE S
Sbjct: 718 LAFEKKD--------SKERHFCLEWLDKQDPNSVLYVSFGTTTTFKEEQIKKIATGLEQS 873
Query: 305 EVPFIWVVRKSAKSE---GENLEW--LPEGFEERIEGSGKGLIIRGWAPQVMILDHESVG 359
+ FIWV+R + K + G +W FEER+EG G L++R WAPQ+ IL H S G
Sbjct: 874 KQKFIWVLRDADKGDIFDGSEAKWNEFSNEFEERVEGMG--LVVRDWAPQLEILSHTSTG 1047
Query: 360 GFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGEP 419
GF++HCGWNS LE +S G+P+ WPM+ +Q N+ +++++KI G+ V+ W
Sbjct: 1048GFMSHCGWNSCLESISMGVPIAAWPMHSDQPRNSVLITEVLKI--GLVVKNWAQRNA--L 1215
Query: 420 VKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLI 470
V +E AVRR+M E ++MR RA + R+++ GG S + + I
Sbjct: 1216VSASNVENAVRRLMETKEGDDMRERAVRLKNVIHRSMDEGGVSRMEIDSFI 1368
>NP600052 zeatin O-glucosyltransferase [Glycine max]
Length = 1395
Score = 201 bits (510), Expect = 8e-52
Identities = 153/482 (31%), Positives = 224/482 (45%), Gaps = 18/482 (3%)
Frame = +1
Query: 7 ELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKINIKTI-- 64
+ ++ PF A GH+ + LAR S + V V T ++ T+ NI I
Sbjct: 31 QTQVVLIPFPAQGHLNQLLHLARHIFSHNIPVHYVGTATHIR--QATLRDHNSNISNIII 204
Query: 65 ---KFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLE----QEKPDCLV 117
F P P N E+ P + K++ LREP+ ++L+ Q K ++
Sbjct: 205 HFHAFEVPPFVSPPPNPNNEETDF-PSHLLPSFKASSHLREPVRNLLQSLSSQAKRVIVI 381
Query: 118 ADMFFPWSTDSAAKF-NIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPG 176
D A N+ FH F + +P V + + +P++ G
Sbjct: 382 HDSLMASVAQDATNMPNVENYTFHSTCAFTTFLYYWEVMGRPP--VEGFFQATEIPSMGG 555
Query: 177 EITLTKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKA 236
PQ F + E E + G I NT +E Y + G K
Sbjct: 556 CFP---------PQ----FIHFITEEYEFHQFNDGNIYNTSRAIEGPYIEFLERIGGSKK 696
Query: 237 --WHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQL 294
W LGP + + ++ R H C++WL +E NSV+YV FG+ T F+ AQ
Sbjct: 697 RLWALGPFNPLTIEKKDPKTR--------HICIEWLDKQEANSVMYVSFGTTTSFTVAQF 852
Query: 295 KEIAMGLEASEVPFIWVVRKSAK------SEGENLEWLPEGFEERIEGSGKGLIIRGWAP 348
++IA+GLE S+ FIWV+R + K SE E E LP GFEER+EG G L+IR WAP
Sbjct: 853 EQIAIGLEQSKQKFIWVLRDADKGNIFDGSEAERYE-LPNGFEERVEGIG--LLIRDWAP 1023
Query: 349 QVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGV 408
Q+ IL H S GGF++HCGWNS LE ++ G+P+ WPM+ +Q N+ +++++K VG V
Sbjct: 1024QLEILSHTSTGGFMSHCGWNSCLESITMGVPIAAWPMHSDQPRNSVLITEVLK--VGFVV 1197
Query: 409 QTWIGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSN 468
+ W V V+E AVRR+M E +EMR RA R+ GG S + +
Sbjct: 1198KDWAQRNA--LVSASVVENAVRRLMETKEGDEMRDRAVRLKNCIHRSKYGGGVSRMEMGS 1371
Query: 469 LI 470
I
Sbjct: 1372FI 1377
>TC227846 weakly similar to UP|Q947K4 (Q947K4) Thiohydroximate
S-glucosyltransferase, partial (49%)
Length = 1605
Score = 193 bits (490), Expect = 2e-49
Identities = 148/474 (31%), Positives = 229/474 (48%), Gaps = 11/474 (2%)
Frame = +2
Query: 11 IFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKINIKTIKFPSPE 70
+ P+ A GHI P V A+ +S+G+K T+ TTH + +I I ++ I +
Sbjct: 2 LVLPYPAQGHINPLVQFAKRLASKGVKATVATTHYT----ANSINAPNITVEAIS-DGFD 166
Query: 71 ETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDCLVADMFFPWSTDSAA 130
+ G + N + LA + + L+R+ H C+V D FFPW D A
Sbjct: 167 QAGFAQTNNNVQLFLASFRTNGSRTLSELIRK---HQQTPSPSTCIVYDSFFPWVLDVAK 337
Query: 131 KFNIPRIVFHGLGFFP----LCVLACTRQYK-PQDKVSSYTEPFVVPNLPGEITLTKMQL 185
+ I +G FF +C + C + Q V P VP LP +
Sbjct: 338 QHGI-----YGAAFFTNSAAVCNIFCRLHHGFIQLPVKMEHLPLRVPGLPPLDSRALPSF 502
Query: 186 PQLPQHDKVFT--KLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWHLGP-V 242
+ P+ + KL + SN + + NTF LE K +GP V
Sbjct: 503 VRFPESYPAYMAMKLSQFSNLNNAD--WMFVNTFEALESEVLKGLTELFPAKM--IGPMV 670
Query: 243 SLCNRDTEEKACRGREASIDE---HECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAM 299
D K +G AS+ + EC WL+SK P SV+Y+ FGSM ++ Q++E+A
Sbjct: 671 PSGYLDGRIKGDKGYGASLWKPLTEECSNWLESKPPQSVVYISFGSMVSLTEEQMEEVAW 850
Query: 300 GLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVG 359
GL+ S V F+WV+R+S + LP G+ E ++ KGLI+ W Q+ +L H++ G
Sbjct: 851 GLKESGVSFLWVLRESEHGK------LPLGYRESVKD--KGLIVT-WCNQLELLAHQATG 1003
Query: 360 GFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGEP 419
FVTHCGWNSTLE +S G+P+V P + +Q +AKFL +I ++GV W
Sbjct: 1004CFVTHCGWNSTLESLSLGVPVVCLPQWADQLPDAKFLDEIWEVGV------WPKEDEKGI 1165
Query: 420 VKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDL 473
V+K ++++ +M G ++E+R A ++ K+AR AV GGSS + ++ L
Sbjct: 1166VRKQEFVQSLKDVMEGQRSQEIRRNANKWKKLAREAVGEGGSSDKHINQFVDHL 1327
>BI970958
Length = 789
Score = 178 bits (452), Expect = 4e-45
Identities = 92/230 (40%), Positives = 142/230 (61%), Gaps = 6/230 (2%)
Frame = -2
Query: 251 EKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEASEVPFIW 310
+K + E +D+ ECLKWL +KE NSV+Y+C GS+ Q EIA G+EAS F+W
Sbjct: 785 DKRGKXXEDXVDD-ECLKWLNTKESNSVVYICXGSLARLXKEQNXEIARGIEASGHKFLW 609
Query: 311 VVRKSAKSEGENLE--WLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGGFVTHCGWN 368
V+ K+ K + E LP GFEER+ +G+++RGW PQ +IL H+++GGF+THCG N
Sbjct: 608 VLPKNTKDDDVKEEELLLPHGFEERMREKKRGMVVRGWVPQGLILKHDAIGGFLTHCGAN 429
Query: 369 STLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGEPVKKDV---- 424
S +E + G+P++T P +G+ F K ++++ +GV +GV W M + K+ V
Sbjct: 428 SVVEAICEGVPLITMPRFGDHFLCEKQATEVLGLGVELGVSEW-SMSPYDARKEVVGWER 252
Query: 425 IEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDLK 474
IE AVR++M DE + R KE + A V+ GG+SY++ + L++ L+
Sbjct: 251 IENAVRKVM-KDEGGLLNKRVKEMKEKAHEVVQEGGNSYDNVTTLVQSLR 105
>TC230019 weakly similar to UP|Q6WFW1 (Q6WFW1) Glucosyltransferase-like
protein , partial (36%)
Length = 1202
Score = 178 bits (452), Expect = 4e-45
Identities = 103/271 (38%), Positives = 154/271 (56%), Gaps = 4/271 (1%)
Frame = +2
Query: 207 VKSFGVIANTFYELEPVYADHYRNELGRKAWHLGPVSLCNRDTEEKACRGREASIDEHEC 266
+KS G I NT E+EP+ RN L W +GP+ + K G+E+ I C
Sbjct: 308 MKSDGWICNTVQEIEPLGLQLLRNYLQLPVWPVGPLLPPASLMDSKHRAGKESGIALDAC 487
Query: 267 LKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVRKSAKSE--GENL- 323
++WL SK+ +SV+Y+ FGS + +Q+ +A GLE S FIW++R + GE +
Sbjct: 488 MQWLDSKDESSVLYISFGSQNTITASQMMALAEGLEESGRSFIWIIRPPFGFDINGEFIA 667
Query: 324 EWLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTW 383
EWLP+GFEER+ + +GL++ W PQ+ IL H S G F++HCGWNS LE +S G+PM+ W
Sbjct: 668 EWLPKGFEERMRDTKRGLLVNKWGPQLEILSHSSTGAFLSHCGWNSVLESLSYGVPMIGW 847
Query: 384 PMYGEQFYNAKFLSDIVKIGVGVGV-QTWIGMGGGEPVKKDVIEKAVRRIMVGDEAEEMR 442
P+ EQ YN K L + ++GV + + QT + G+ VKK VIE A+ + + +EM+
Sbjct: 848 PLAAEQTYNLKML--VEEMGVAIELTQTVETVISGKQVKK-VIEIAMEQ---EGKGKEMK 1009
Query: 443 SRAKEFGKMARRAVEVGGSSYNDFSNLIEDL 473
+A E R A+ G ++DL
Sbjct: 1010EKANEIAAHMREAITEKGKEKGSSVRAMDDL 1102
>TC219585 weakly similar to UP|Q8L5C7 (Q8L5C7) UDP-glucuronosyltransferase ,
partial (50%)
Length = 935
Score = 174 bits (442), Expect = 6e-44
Identities = 88/213 (41%), Positives = 132/213 (61%), Gaps = 1/213 (0%)
Frame = +2
Query: 262 DEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVRKSAKSEGE 321
++ +C++WL EP+SVIYV +GS+TV S+ LKE A GL S +PF+W+ R GE
Sbjct: 74 NDSKCIQWLDQWEPSSVIYVNYGSITVMSEDHLKEFAWGLANSNLPFLWIKRPDLVM-GE 250
Query: 322 NLEWLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMV 381
+ + LP+ F + ++ G I GW PQ +L H SVG F+THCGWNSTLEG+S G+PM+
Sbjct: 251 STQ-LPQDFLDEVKDRG---YITGWCPQEQVLSHPSVGVFLTHCGWNSTLEGISGGVPMI 418
Query: 382 TWPMYGEQFYNAKFLSDIVKIGVGVGVQTW-IGMGGGEPVKKDVIEKAVRRIMVGDEAEE 440
WP + EQ N +++ TW IGM + VK++ + V+ ++ G+ +E
Sbjct: 419 GWPFFAEQQTNCRYI-----------CTTWGIGMDIKDDVKREEVTTLVKEMITGERGKE 565
Query: 441 MRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDL 473
MR + E+ K A A ++GGSSYNDF L++++
Sbjct: 566 MRQKCLEWKKKAIEATDMGGSSYNDFHRLVKEV 664
>TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (Fragment),
partial (58%)
Length = 1530
Score = 174 bits (442), Expect = 6e-44
Identities = 147/491 (29%), Positives = 229/491 (45%), Gaps = 25/491 (5%)
Frame = +1
Query: 9 HIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLN--VPLISRTIGKAKINIKTIKF 66
H+ P H++P ++ FS R L + H+ +P + + +K ++T+
Sbjct: 4 HVAVVPSPGFTHLVPILE----FSKRLLHLH-PEFHITCFIPSVGSSPTSSKAYVQTLP- 165
Query: 67 PSPEETGLPE---GCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDCLVADMFFP 123
P+ LP + S LA + S +RE L+ + + K LV D+F
Sbjct: 166 PTITSIFLPPITLDHVSDPSVLALQIELSVNLSLPYIREELKSLCSRAKVVALVVDVFAN 345
Query: 124 WSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQY---KPQDKVSSYTEPFVVP-NLPGEIT 179
+ + A + N+ ++ P + + + K + +SS + P ++PG +
Sbjct: 346 GALNFAKELNLLSYIY-----LPQSAMLLSLYFYSTKLDEILSSESRELQKPIDIPGCVP 510
Query: 180 LTKMQLPQLPQHDKV---FTKLLEESNESEVKSFGVIANTFYELEP----VYADHYRNEL 232
+ LP LP HD + LE S V GV NTF ELE +H + +
Sbjct: 511 IHNKDLP-LPFHDLSGLGYKGFLERSKRFHVPD-GVFMNTFLELESGAIRALEEHVKGK- 681
Query: 233 GRKAWHLGPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDA 292
K + +GP+ + G E + ECL WL +EPNSV+YV FGS S
Sbjct: 682 -PKLYPVGPII-------QMESIGHENGV---ECLTWLDKQEPNSVLYVSFGSGGTLSQE 828
Query: 293 QLKEIAMGLEASEVPFIWVVR--KSAKSEG-------ENLEWLPEGFEERIEGSGKGLII 343
Q E+A GLE S F+WVVR S G + LE+LP GF ER + +GL++
Sbjct: 829 QFNELAFGLELSGKKFLWVVRAPSGVVSAGYLCAETKDPLEFLPHGFLERTKK--QGLVV 1002
Query: 344 RGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIG 403
WAPQ+ +L H + GGF++HCGWNS LE V G+P++TWP++ EQ NA ++D +K+
Sbjct: 1003PSWAPQIQVLGHSATGGFLSHCGWNSVLESVVQGVPVITWPLFAEQSLNAAMIADDLKVA 1182
Query: 404 VGVGVQTWIGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSY 463
+ V V+++ I K VR +M E+ E+R R A A++ GSS
Sbjct: 1183LRPKVNE------SGLVEREEIAKVVRGLMGDKESLEIRKRMGLLKIAAANAIKEDGSST 1344
Query: 464 NDFSNLIEDLK 474
S + L+
Sbjct: 1345KTLSEMATSLR 1377
>TC231296 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (41%)
Length = 808
Score = 173 bits (439), Expect = 1e-43
Identities = 94/281 (33%), Positives = 154/281 (54%), Gaps = 1/281 (0%)
Frame = +1
Query: 1 MTNENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKIN 60
M + + L + F P+L+ GH+IP +A +F+SRG VT++TT ++ ++ +++
Sbjct: 13 MDLQQRPLKLHFIPYLSPGHVIPLCGIATLFASRGQHVTVITTPYYAQILRKSSPSLQLH 192
Query: 61 IKTIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDCLVADM 120
+ + FP+ ++ GLP+G E + KF ++ +LLR P+ H ++Q PDC+VAD
Sbjct: 193 V--VDFPA-KDVGLPDGVEIKSAVTDLADTAKFYQAAMLLRRPISHFMDQHPPDCIVADT 363
Query: 121 FFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEITL 180
+ W+ D A IPR+ F+G F + C + ++ S T PFV+P+ P +T
Sbjct: 364 MYSWADDVANNLRIPRLAFNGYPLFSGAAMKCVISH---PELHSDTGPFVIPDFPHRVT- 531
Query: 181 TKMQLPQLPQHDKVFTKLLEESNESEVKSFGVIANTFYELE-PVYADHYRNELGRKAWHL 239
+P P K+ T ++ + E+KS G+I N+F EL+ HY G KAWHL
Sbjct: 532 ----MPSRP--PKMATAFMDHLLKIELKSHGLIVNSFAELDGEECIQHYEKSTGHKAWHL 693
Query: 240 GPVSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSVIY 280
GP L + +E RG ++ + ++ECL WL K NSV+Y
Sbjct: 694 GPACLVGKRDQE---RGEKSVVSQNECLTWLDPKPTNSVVY 807
>BM187903
Length = 438
Score = 172 bits (436), Expect = 3e-43
Identities = 81/144 (56%), Positives = 106/144 (73%), Gaps = 5/144 (3%)
Frame = +3
Query: 269 WLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVRKSAKSEGENLE---- 324
WL SK+PNSV+YV FGS+ QLKEIA GLEAS+ FIWVV + EN E
Sbjct: 6 WLNSKKPNSVLYVSFGSVARLPPGQLKEIAFGLEASDQTFIWVVGCIRNNPSENKENGSG 185
Query: 325 -WLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTW 383
+L EGFE+R++ KGL++RGWAPQ++IL+H ++ GF+THCGWNSTLE V AG+PM+TW
Sbjct: 186 NFLFEGFEQRMKEKNKGLVLRGWAPQLLILEHAAIKGFMTHCGWNSTLESVCAGVPMITW 365
Query: 384 PMYGEQFYNAKFLSDIVKIGVGVG 407
P+ EQF N K +++++KIGV VG
Sbjct: 366 PLSAEQFSNEKLITEVLKIGVQVG 437
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,806,439
Number of Sequences: 63676
Number of extensions: 322152
Number of successful extensions: 1826
Number of sequences better than 10.0: 196
Number of HSP's better than 10.0 without gapping: 1638
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1659
length of query: 478
length of database: 12,639,632
effective HSP length: 101
effective length of query: 377
effective length of database: 6,208,356
effective search space: 2340550212
effective search space used: 2340550212
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC136449.6


