

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136287.2 + phase: 0
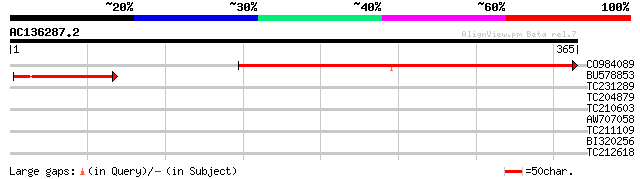
(365 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CO984089 357 5e-99
BU578853 100 8e-22
TC231289 30 1.3
TC204879 similar to UP|Q8RX88 (Q8RX88) AT4g01320/F2N1_21, partia... 29 2.9
TC210603 similar to UP|Q9LHJ9 (Q9LHJ9) Protein phosphatase 2C (A... 28 5.0
AW707058 similar to GP|18377851|gb| AT3g56680/T8M16_10 {Arabidop... 28 5.0
TC211109 weakly similar to UP|O48631 (O48631) Ethylene-forming-e... 28 6.5
BI320256 28 8.5
TC212618 weakly similar to UP|Q9FZI1 (Q9FZI1) F1O19.5 protein, p... 28 8.5
>CO984089
Length = 827
Score = 357 bits (916), Expect = 5e-99
Identities = 185/220 (84%), Positives = 197/220 (89%), Gaps = 2/220 (0%)
Frame = -1
Query: 148 HGLDGIVLKVEDVEPMLELKEYFDRRTEESNVLNLTKATVTQIQVAGMGDRVCVDLCSLM 207
HGLDGI++KVEDVE +LELKEYFDRR EESN+L+LTKATVT IQ AGMGDRVCVDLCSLM
Sbjct: 827 HGLDGIIMKVEDVEXVLELKEYFDRRMEESNLLSLTKATVTHIQAAGMGDRVCVDLCSLM 648
Query: 208 RPGEGLLVGSFARGLFLVHSECLESKLHCKQTFSGEC--VHAYIAVPGGRTSYLSELKSG 265
RPGEGLLVGSFARGLFLVHSECLES + F VHAY+AVPGGRT YLSELKSG
Sbjct: 647 RPGEGLLVGSFARGLFLVHSECLESNYIASRPFRVNAGPVHAYVAVPGGRTCYLSELKSG 468
Query: 266 KEVIVVDQQGRQRIAIVGRVKIESRPLILVEAKTDSDNQTISILLQNAETVALVCPVQGN 325
KEVI+VD QGRQRIAIVGRVKIESRPLILVEAK +SDNQ+ISILLQNAETVALVC QGN
Sbjct: 467 KEVIIVDHQGRQRIAIVGRVKIESRPLILVEAKIESDNQSISILLQNAETVALVCTPQGN 288
Query: 326 KLLKTAFPVTSLKVGDEVLLRVQGGARHTGIEIQEFIVEK 365
LLKT+ PVTSLKVGDE+LLRVQGGARHTGIEIQEFIVEK
Sbjct: 287 TLLKTSIPVTSLKVGDEILLRVQGGARHTGIEIQEFIVEK 168
>BU578853
Length = 451
Score = 100 bits (250), Expect = 8e-22
Identities = 48/67 (71%), Positives = 57/67 (84%)
Frame = +2
Query: 3 SSSWTPLEDSGNKKSKKVWIWTKNKQVMTAAVERGWNTFIFPSNLPHLANDWSSIAVICP 62
SS + L++SG K+SK+VWIWT NKQVMTAAVERGWNTF+FPS+ LA+DWSSIAVICP
Sbjct: 254 SSMASSLDESG-KRSKRVWIWTSNKQVMTAAVERGWNTFVFPSHHRQLAHDWSSIAVICP 430
Query: 63 LFASEKE 69
LF +E E
Sbjct: 431 LFVNEGE 451
>TC231289
Length = 694
Score = 30.4 bits (67), Expect = 1.3
Identities = 17/39 (43%), Positives = 21/39 (53%)
Frame = +1
Query: 214 LVGSFARGLFLVHSECLESKLHCKQTFSGECVHAYIAVP 252
L+ S ++ LF CL S LH S CVHA IA+P
Sbjct: 163 LIASSSQDLF---GRCLMSSLHIFSPNSNLCVHAAIAIP 270
>TC204879 similar to UP|Q8RX88 (Q8RX88) AT4g01320/F2N1_21, partial (56%)
Length = 1057
Score = 29.3 bits (64), Expect = 2.9
Identities = 22/70 (31%), Positives = 33/70 (46%), Gaps = 1/70 (1%)
Frame = -1
Query: 229 CLESKL-HCKQTFSGECVHAYIAVPGGRTSYLSELKSGKEVIVVDQQGRQRIAIVGRVKI 287
C+ SKL CK S +CVH+ + +P S L S +IV A++ I
Sbjct: 388 CISSKL*KCKNLHSNKCVHSMVELP---VSQLMGNNSNNFLIV--------FALLN*CVI 242
Query: 288 ESRPLILVEA 297
+ PL+L E+
Sbjct: 241 KDNPLVLEES 212
>TC210603 similar to UP|Q9LHJ9 (Q9LHJ9) Protein phosphatase 2C (At3g12620),
partial (75%)
Length = 883
Score = 28.5 bits (62), Expect = 5.0
Identities = 22/64 (34%), Positives = 29/64 (44%)
Frame = +2
Query: 49 HLANDWSSIAVICPLFASEKEILDAQNKKVATIFDVSTPEELERLRPEDEQAENIVVNLL 108
++AN S AV+ L + KEI D Q S EEL L P D Q +V+
Sbjct: 305 YIANAGDSRAVLGRLDEAMKEIKDIQLSVEHNASHASVREELHSLHPNDPQI--VVMKHQ 478
Query: 109 DWQV 112
W+V
Sbjct: 479 VWRV 490
>AW707058 similar to GP|18377851|gb| AT3g56680/T8M16_10 {Arabidopsis
thaliana}, partial (5%)
Length = 352
Score = 28.5 bits (62), Expect = 5.0
Identities = 12/47 (25%), Positives = 26/47 (54%)
Frame = +2
Query: 266 KEVIVVDQQGRQRIAIVGRVKIESRPLILVEAKTDSDNQTISILLQN 312
K++I+ + + + VKI+ PLI ++ + + NQT +++ N
Sbjct: 179 KKIIIRTRPNKNNLNNTNEVKIKKNPLISIK*RKEKYNQTQTLIFSN 319
>TC211109 weakly similar to UP|O48631 (O48631) Ethylene-forming-enzyme-like
dioxygenase, partial (48%)
Length = 774
Score = 28.1 bits (61), Expect = 6.5
Identities = 21/82 (25%), Positives = 38/82 (45%), Gaps = 3/82 (3%)
Frame = +3
Query: 271 VDQQGRQRIAIV--GRVKIESRPLILVEAKTDSDNQTISILLQNAETVALVCPVQGNKLL 328
VDQ G Q + + + SRP +++ K +D I++LLQ+ E L + N +
Sbjct: 156 VDQFGEQPLMLARFNFYPLCSRPDLVLGVKPHTDRSGITVLLQDKEVEGLQVLIDDNWIN 335
Query: 329 KTAFP-VTSLKVGDEVLLRVQG 349
P + +GD++ + G
Sbjct: 336 VPTMPDALVVNLGDQMQIMSNG 401
>BI320256
Length = 109
Score = 27.7 bits (60), Expect = 8.5
Identities = 11/18 (61%), Positives = 13/18 (72%)
Frame = +1
Query: 219 ARGLFLVHSECLESKLHC 236
A+GL +H ECLE LHC
Sbjct: 55 AKGLAYLHEECLEWILHC 108
>TC212618 weakly similar to UP|Q9FZI1 (Q9FZI1) F1O19.5 protein, partial (13%)
Length = 367
Score = 27.7 bits (60), Expect = 8.5
Identities = 12/46 (26%), Positives = 23/46 (49%), Gaps = 5/46 (10%)
Frame = +2
Query: 4 SSWTPLEDSGNKKSKKVW-----IWTKNKQVMTAAVERGWNTFIFP 44
S+W D K +W +W+K ++ A+++GW+ +FP
Sbjct: 80 STWWVFRDISRFKFTCLWFGT*YLWSKCDSMV*CAIDKGWSWDLFP 217
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.133 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,176,293
Number of Sequences: 63676
Number of extensions: 176794
Number of successful extensions: 872
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 869
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 871
length of query: 365
length of database: 12,639,632
effective HSP length: 99
effective length of query: 266
effective length of database: 6,335,708
effective search space: 1685298328
effective search space used: 1685298328
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC136287.2


