

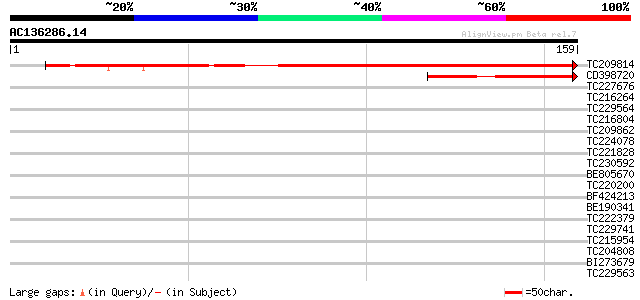
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136286.14 - phase: 0
(159 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC209814 142 5e-35
CD398720 60 3e-10
TC227676 39 0.001
TC216264 similar to UP|Q9MA01 (Q9MA01) F20B17.20, partial (10%) 37 0.005
TC229564 weakly similar to UP|Q6VSS7 (Q6VSS7) TOX, partial (18%) 35 0.015
TC216804 similar to UP|PFD5_ARATH (P57742) Probable prefoldin su... 34 0.026
TC209862 similar to PIR|C86255|C86255 protein F12F1.11 [imported... 34 0.026
TC224078 similar to UP|Q75DD0 (Q75DD0) ABR097Cp, partial (4%) 34 0.026
TC221828 similar to PIR|T02573|T02573 CDP-diacylglycerol-glycero... 34 0.034
TC230592 similar to UP|Q902U4 (Q902U4) Rev protein, partial (16%) 33 0.058
BE805670 32 0.100
TC220200 weakly similar to UP|Q7M4A3 (Q7M4A3) Spermatid-specific... 32 0.13
BF424213 weakly similar to GP|21322711|em pherophorin-dz1 protei... 32 0.17
BE190341 32 0.17
TC222379 homologue to UP|Q9HGV7 (Q9HGV7) Gbeta like protein, par... 32 0.17
TC229741 32 0.17
TC215954 similar to UP|Q9LNB6 (Q9LNB6) F5O11.2 (At1g12270/F5O11_... 31 0.22
TC204808 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone g... 31 0.22
BI273679 31 0.29
TC229563 30 0.38
>TC209814
Length = 639
Score = 142 bits (359), Expect = 5e-35
Identities = 88/154 (57%), Positives = 103/154 (66%), Gaps = 5/154 (3%)
Frame = +1
Query: 11 LTLSPHFTPSPPSTLS-PQFFNFKLKP----SLPSLTLTRRCTLPRAVEEKDQPTSNPEP 65
LT HF PSPP L+ FK +P + L RR + AVEE Q +S PE
Sbjct: 28 LTSQSHF-PSPPLRLTLTASAPFKPRPLASIGISPLPERRRMPVAGAVEES-QESSEPEA 201
Query: 66 EPEPKPELNESELASELKKAMQERKEQEGNNFWNGVVSEIGEIEWPEFGKVLGTTGVVLS 125
E ++LASELKKAM ERKE+E N +GV EI EIEWP FGKVLGTTGVVLS
Sbjct: 202 E---------ADLASELKKAMAERKEKEEENMLSGVAREIDEIEWPAFGKVLGTTGVVLS 354
Query: 126 VIFGSSAVLLTLNAILAELSDKVFAGKGVQDFFT 159
VIFGSS VLL++NA+LAE+SD+VFAGKG+QDFFT
Sbjct: 355 VIFGSSVVLLSVNAVLAEISDRVFAGKGIQDFFT 456
>CD398720
Length = 684
Score = 60.5 bits (145), Expect = 3e-10
Identities = 31/42 (73%), Positives = 36/42 (84%)
Frame = +3
Query: 118 GTTGVVLSVIFGSSAVLLTLNAILAELSDKVFAGKGVQDFFT 159
GTTG VLSVIFGSS +NA+LA+LSD+VFAGKG+QDFFT
Sbjct: 297 GTTGDVLSVIFGSS-----VNAVLADLSDRVFAGKGIQDFFT 407
>TC227676
Length = 1104
Score = 38.9 bits (89), Expect = 0.001
Identities = 34/92 (36%), Positives = 45/92 (47%), Gaps = 6/92 (6%)
Frame = +1
Query: 2 VLQAKTSFTLTLSPHFTPSPPST--LSPQFFNFKLKPS----LPSLTLTRRCTLPRAVEE 55
+LQ TSF L+L H P+PP T L P+ KP PSL R ++ +
Sbjct: 256 LLQRVTSFNLSLHKH-QPTPPETQHLHPEPQPQPEKPPELLRTPSLLERLRSFHNLSLHK 432
Query: 56 KDQPTSNPEPEPEPKPELNESELASELKKAMQ 87
QP PEPEPEP+PE E + L + +Q
Sbjct: 433 HVQP--EPEPEPEPEPEKPELVRSPSLLQRIQ 522
>TC216264 similar to UP|Q9MA01 (Q9MA01) F20B17.20, partial (10%)
Length = 1332
Score = 36.6 bits (83), Expect = 0.005
Identities = 23/62 (37%), Positives = 29/62 (46%), Gaps = 2/62 (3%)
Frame = -3
Query: 12 TLSPHFTPSPPSTLSPQFFNFKLKP-SLPSLT-LTRRCTLPRAVEEKDQPTSNPEPEPEP 69
+LSP PP+T+ + + P S PS TR CT P A PT+ P P P P
Sbjct: 511 SLSPSLPSQPPNTV-----HISIDPRSAPSRAQCTRACTPPSARSSSTSPTTPPSPPPPP 347
Query: 70 KP 71
P
Sbjct: 346 PP 341
>TC229564 weakly similar to UP|Q6VSS7 (Q6VSS7) TOX, partial (18%)
Length = 609
Score = 35.0 bits (79), Expect = 0.015
Identities = 16/43 (37%), Positives = 26/43 (60%)
Frame = +3
Query: 51 RAVEEKDQPTSNPEPEPEPKPELNESELASELKKAMQERKEQE 93
R +++ + PEPEPE K E E+E +E + +E+KE+E
Sbjct: 78 RRTKKQAKIVPQPEPEPEKKEESKEAEKPAEEENKPEEKKEEE 206
>TC216804 similar to UP|PFD5_ARATH (P57742) Probable prefoldin subunit 5,
partial (85%)
Length = 923
Score = 34.3 bits (77), Expect = 0.026
Identities = 24/85 (28%), Positives = 38/85 (44%)
Frame = +3
Query: 6 KTSFTLTLSPHFTPSPPSTLSPQFFNFKLKPSLPSLTLTRRCTLPRAVEEKDQPTSNPEP 65
K F+ T SP F P PP++ SP + + PS P T + + P P+++P P
Sbjct: 291 KLIFSKTASPTFAPPPPASRSPPPPS-TISPSAPRATTSSSPSPP--------PSTSPPP 443
Query: 66 EPEPKPELNESELASELKKAMQERK 90
P + S A+ +K + K
Sbjct: 444 STTPNTSSSTSAPATSSRKPCPKEK 518
>TC209862 similar to PIR|C86255|C86255 protein F12F1.11 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(40%)
Length = 933
Score = 34.3 bits (77), Expect = 0.026
Identities = 25/60 (41%), Positives = 29/60 (47%)
Frame = +2
Query: 12 TLSPHFTPSPPSTLSPQFFNFKLKPSLPSLTLTRRCTLPRAVEEKDQPTSNPEPEPEPKP 71
T+ FT PS SP +PS PS RR + PR EK PTS+P P P P P
Sbjct: 38 TIKGRFTRRHPS--SPTNSPSSRRPSSPS----RRRSHPRT--EKSSPTSSPAPPPPPPP 193
>TC224078 similar to UP|Q75DD0 (Q75DD0) ABR097Cp, partial (4%)
Length = 364
Score = 34.3 bits (77), Expect = 0.026
Identities = 19/57 (33%), Positives = 30/57 (52%), Gaps = 1/57 (1%)
Frame = +2
Query: 22 PSTLSPQFFNFKLKPSL-PSLTLTRRCTLPRAVEEKDQPTSNPEPEPEPKPELNESE 77
P L+P F F P L P+L P + K P+ NP P+P+PKP++ +++
Sbjct: 59 PPFLNPNFLCFN--PFLNPNLLCFNPFLHPNNLPPKALPSLNPNPKPDPKPQIEKTQ 223
>TC221828 similar to PIR|T02573|T02573
CDP-diacylglycerol-glycerol-3-phosphate
3-phosphatidyltransferase homolog At2g39290 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(22%)
Length = 437
Score = 33.9 bits (76), Expect = 0.034
Identities = 27/84 (32%), Positives = 35/84 (41%), Gaps = 7/84 (8%)
Frame = +1
Query: 2 VLQAKTSFTLTLSPHFTPSPPST----LSP---QFFNFKLKPSLPSLTLTRRCTLPRAVE 54
V AKT LS P P + LSP Q F + P +L R +
Sbjct: 1 VRTAKTKSKPLLSWRSPPQHPRSRRYMLSPSKNQILGFTPRNKFPCYSLGRAGLVTDFGS 180
Query: 55 EKDQPTSNPEPEPEPKPELNESEL 78
QP P+P+PEP+PE S+L
Sbjct: 181SVPQPQPQPQPKPEPEPEPEPSKL 252
>TC230592 similar to UP|Q902U4 (Q902U4) Rev protein, partial (16%)
Length = 1282
Score = 33.1 bits (74), Expect = 0.058
Identities = 21/66 (31%), Positives = 29/66 (43%), Gaps = 9/66 (13%)
Frame = +1
Query: 19 PSPPSTLS------PQFFNFKLKP---SLPSLTLTRRCTLPRAVEEKDQPTSNPEPEPEP 69
PSPP T S P+ F+ P S P +L + P+ + NP P P P
Sbjct: 34 PSPPKTTSFSSLPQPKSSLFQSLPQPKSSPFSSLFQSLPPPKQPSSESASLPNPNPNPNP 213
Query: 70 KPELNE 75
KP++ E
Sbjct: 214KPQIEE 231
>BE805670
Length = 416
Score = 32.3 bits (72), Expect = 0.100
Identities = 20/57 (35%), Positives = 27/57 (47%), Gaps = 1/57 (1%)
Frame = +2
Query: 14 SPHFTPSPPSTLSPQFFNFKLKPSLPSLTLTRRC-TLPRAVEEKDQPTSNPEPEPEP 69
+P + PSP S +P L P S + TRR P+ V + +P SNP P P
Sbjct: 236 TPQWLPSPASRKTP--LRPSLAPMCSSTSTTRRS*WKPKVVAPRSKPYSNPPVSPSP 400
>TC220200 weakly similar to UP|Q7M4A3 (Q7M4A3) Spermatid-specific protein T2
precursor, partial (40%)
Length = 506
Score = 32.0 bits (71), Expect = 0.13
Identities = 22/69 (31%), Positives = 29/69 (41%), Gaps = 2/69 (2%)
Frame = +2
Query: 5 AKTSFTLTLSPHFTPSP--PSTLSPQFFNFKLKPSLPSLTLTRRCTLPRAVEEKDQPTSN 62
A T+ T SP TP+P P + SP F + PS + + P+SN
Sbjct: 185 AATAATAAESPAPTPTPDFPPSSSPPFPSPASNPSTANFPPSPTPAPSHHSPPAPSPSSN 364
Query: 63 PEPEPEPKP 71
P P P P P
Sbjct: 365 PSPSPAPAP 391
>BF424213 weakly similar to GP|21322711|em pherophorin-dz1 protein {Volvox
carteri f. nagariensis}, partial (6%)
Length = 424
Score = 31.6 bits (70), Expect = 0.17
Identities = 29/76 (38%), Positives = 37/76 (48%), Gaps = 5/76 (6%)
Frame = +1
Query: 1 MVLQAKTSFTLTLSPHF---TPS--PPSTLSPQFFNFKLKPSLPSLTLTRRCTLPRAVEE 55
++ +A S + TL P TPS PPS+L+P F L PS P+LT TLP
Sbjct: 118 LLSKAPRSLSHTLPPSLPLHTPSQPPPSSLNPPPF---LLPS-PTLTPPSLSTLPSPPFP 285
Query: 56 KDQPTSNPEPEPEPKP 71
+P P P P P P
Sbjct: 286 LPKP*P*P*PSPPPPP 333
>BE190341
Length = 274
Score = 31.6 bits (70), Expect = 0.17
Identities = 19/51 (37%), Positives = 23/51 (44%)
Frame = -2
Query: 19 PSPPSTLSPQFFNFKLKPSLPSLTLTRRCTLPRAVEEKDQPTSNPEPEPEP 69
P PPS+LS Q S P TLT R ++P + P P P P P
Sbjct: 273 PLPPSSLSSQ--------SRPPSTLTPRHSIPLSTLRASVPRITPSPSPVP 145
>TC222379 homologue to UP|Q9HGV7 (Q9HGV7) Gbeta like protein, partial (84%)
Length = 950
Score = 31.6 bits (70), Expect = 0.17
Identities = 24/67 (35%), Positives = 29/67 (42%), Gaps = 9/67 (13%)
Frame = +2
Query: 10 TLTLSPHFTPSPPSTLSPQFFNFKLKPSLP--SLTLTRRCTL-------PRAVEEKDQPT 60
T TLSP T S PS+ P P LP S TL RR L R+ P+
Sbjct: 524 TSTLSPLVTRSTPSSSPPTATGXAPPPPLPSSSSTLRRRARLTSSSPSTSRSARRPASPS 703
Query: 61 SNPEPEP 67
++P P P
Sbjct: 704 ASPSPGP 724
>TC229741
Length = 610
Score = 31.6 bits (70), Expect = 0.17
Identities = 17/57 (29%), Positives = 25/57 (43%)
Frame = +2
Query: 16 HFTPSPPSTLSPQFFNFKLKPSLPSLTLTRRCTLPRAVEEKDQPTSNPEPEPEPKPE 72
H PP + + ++PS P L + + PT PEPEPEP+P+
Sbjct: 68 HGHEHPPHYCCSCYCSSSIQPSAPPLPSSSCLDFEYSNYGTFLPTPEPEPEPEPQPQ 238
>TC215954 similar to UP|Q9LNB6 (Q9LNB6) F5O11.2 (At1g12270/F5O11_1), partial
(76%)
Length = 1839
Score = 31.2 bits (69), Expect = 0.22
Identities = 17/42 (40%), Positives = 23/42 (54%)
Frame = +3
Query: 54 EEKDQPTSNPEPEPEPKPELNESELASELKKAMQERKEQEGN 95
E QP S PEPEPE EL E ++ +KA +++ GN
Sbjct: 498 EPAKQPESEPEPEPE-SMELTGEEKGAKQRKAEALKEKDAGN 620
>TC204808 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone
glucosyltransferase (Arbutin synthase) , partial (25%)
Length = 431
Score = 31.2 bits (69), Expect = 0.22
Identities = 24/73 (32%), Positives = 30/73 (40%)
Frame = +3
Query: 12 TLSPHFTPSPPSTLSPQFFNFKLKPSLPSLTLTRRCTLPRAVEEKDQPTSNPEPEPEPKP 71
T SP TPSPPS+ + P PS T P QP S+P P P P
Sbjct: 51 TPSPPHTPSPPSSSTS-------SPPTPS-TSPPNSMPPPTSSTPPQPPSSPSPSTSP-P 203
Query: 72 ELNESELASELKK 84
+ S +SE +
Sbjct: 204 STSRSSASSETSR 242
>BI273679
Length = 380
Score = 30.8 bits (68), Expect = 0.29
Identities = 12/23 (52%), Positives = 15/23 (65%)
Frame = -1
Query: 56 KDQPTSNPEPEPEPKPELNESEL 78
+ QP PEPEPEP+PE S +
Sbjct: 74 QSQPGPEPEPEPEPEPEPESSRV 6
Score = 29.3 bits (64), Expect = 0.84
Identities = 11/22 (50%), Positives = 14/22 (63%)
Frame = -1
Query: 51 RAVEEKDQPTSNPEPEPEPKPE 72
R + + P PEPEPEP+PE
Sbjct: 83 RRFQSQPGPEPEPEPEPEPEPE 18
>TC229563
Length = 669
Score = 30.4 bits (67), Expect = 0.38
Identities = 17/45 (37%), Positives = 25/45 (54%), Gaps = 1/45 (2%)
Frame = +1
Query: 51 RAVEEKDQPTSNPEPEPEPKPEL-NESELASELKKAMQERKEQEG 94
R +++ + PEPEPE K E E E E +K +E K++EG
Sbjct: 55 RRTKKQAKIVPQPEPEPEKKEEKPAEEETKPEEEKPPEEAKKEEG 189
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.132 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,177,611
Number of Sequences: 63676
Number of extensions: 117609
Number of successful extensions: 1753
Number of sequences better than 10.0: 213
Number of HSP's better than 10.0 without gapping: 1368
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1592
length of query: 159
length of database: 12,639,632
effective HSP length: 89
effective length of query: 70
effective length of database: 6,972,468
effective search space: 488072760
effective search space used: 488072760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC136286.14


