

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136286.11 - phase: 0
(620 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
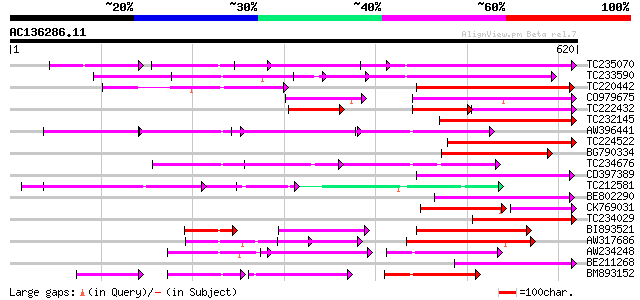
Score E
Sequences producing significant alignments: (bits) Value
TC235070 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding p... 158 8e-39
TC233590 similar to UP|Q6K9T2 (Q6K9T2) Pentatricopeptide repeat-... 156 3e-38
TC220442 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding p... 137 1e-32
CO979675 128 9e-30
TC222432 77 2e-28
TC232145 120 2e-27
AW396441 119 3e-27
TC224522 weakly similar to UP|Q9LUL5 (Q9LUL5) Emb|CAB66100.1, pa... 117 2e-26
BG790334 weakly similar to GP|2160154|gb| F19K23.18 gene product... 114 1e-25
TC234676 114 2e-25
CD397389 similar to GP|6691205|gb| F1K23.11 {Arabidopsis thalian... 114 2e-25
TC212581 weakly similar to UP|Q6K957 (Q6K957) Pentatricopeptide ... 109 3e-24
BE802290 105 5e-23
CK769031 76 1e-22
TC234029 similar to UP|Q9LN01 (Q9LN01) T6D22.15, partial (32%) 103 2e-22
BI893521 103 3e-22
AW317686 102 4e-22
AW234248 102 7e-22
BE211268 100 1e-21
BM893152 98 1e-20
>TC235070 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding protein-like,
partial (22%)
Length = 875
Score = 158 bits (399), Expect = 8e-39
Identities = 91/238 (38%), Positives = 134/238 (56%), Gaps = 2/238 (0%)
Frame = +2
Query: 384 CGDSCVLRSAKAIHGYALRICVDKDDFLATALVDTYMKCGCVSFARFVFDQFDVKPDDPA 443
C C R + +H + ++ + ++ TALVD Y KCG ++ A+ F + + A
Sbjct: 5 CSCLCSFRQGQLLHAHLIKTPFQVNVYVGTALVDFYSKCGHLAEAQRSF--ISIFSPNVA 178
Query: 444 FWNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFFR-M 502
W A+I GY +G A +F ML + + PN+ATFV VLSAC+H+G + GLR F M
Sbjct: 179 AWTALINGYAYHGLGSEAILLFRSMLHQGIVPNAATFVGVLSACNHAGLVCEGLRIFHSM 358
Query: 503 IRKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQEL-AEPPASVFDSLLGACRCYLDSNL 561
R YG+ P EH+ CVVDLLGR+G L EA + + ++ E ++ +LL A + D +
Sbjct: 359 QRCYGVTPTIEHYTCVVDLLGRSGHLKEAEEFIIKMPIEADGIIWGALLNASWFWKDMEV 538
Query: 562 GEEMAMKLIDIEPKNPAPLVVLSNIYAALGRWSEVERIRGLITDKGLDKNSGISMIEV 619
GE A KL ++P VVLSN+YA LGRW + ++R + L K+ G S IE+
Sbjct: 539 GERAAEKLFSLDPNPIFAFVVLSNMYAILGRWGQKTKLRKRLQSLELRKDPGCSWIEL 712
Score = 82.0 bits (201), Expect = 7e-16
Identities = 51/172 (29%), Positives = 89/172 (51%), Gaps = 1/172 (0%)
Frame = +2
Query: 247 ACATLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSEKRNLIT 306
AC+ L + R G+ +H +K +V V T+LVD YSKCG A F N+
Sbjct: 2 ACSCLCSFRQGQLLHAHLIKTPFQVNVYVGTALVDFYSKCGHLAEAQRSFISIFSPNVAA 181
Query: 307 WNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFSKMQ 366
W ++I G + A+ LF M+ +GI+P++AT+ ++S G+ E + F MQ
Sbjct: 182 WTALINGYAYHGLGSEAILLFRSMLHQGIVPNAATFVGVLSACNHAGLVCEGLRIFHSMQ 361
Query: 367 -CAGVAPCLKILTSLLSVCGDSCVLRSAKAIHGYALRICVDKDDFLATALVD 417
C GV P ++ T ++ + G S L+ A+ + +++ ++ D + AL++
Sbjct: 362 RCYGVTPTIEHYTCVVDLLGRSGHLKEAEE---FIIKMPIEADGIIWGALLN 508
Score = 62.0 bits (149), Expect = 8e-10
Identities = 38/103 (36%), Positives = 56/103 (53%)
Frame = +2
Query: 44 ACSNLSSPSQTQILHAHLFKTGFHSHPHTSTALIASYAANTRSFHYALELFDEMPQPTIT 103
ACS L S Q Q+LHAHL KT F + + TAL+ Y + A F + P +
Sbjct: 2 ACSCLCSFRQGQLLHAHLIKTPFQVNVYVGTALVDFY-SKCGHLAEAQRSFISIFSPNVA 178
Query: 104 AFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPNSVTIVSLLSA 146
A+ A+++G + +G +A+ LFR + I PN+ T V +LSA
Sbjct: 179 AWTALINGYAYHGLGSEAILLFRSMLHQGIVPNAATFVGVLSA 307
Score = 55.1 bits (131), Expect = 9e-08
Identities = 37/133 (27%), Positives = 62/133 (45%), Gaps = 1/133 (0%)
Frame = +2
Query: 156 QQVHCLACKLGVEYDVYVSTSLVTAYSKCGVLVSSNKVFENLRVKNVVTYNAFMSGLLQN 215
Q +H K + +VYV T+LV YSKCG L + + F ++ NV + A ++G +
Sbjct: 35 QLLHAHLIKTPFQVNVYVGTALVDFYSKCGHLAEAQRSFISIFSPNVAAWTALINGYAYH 214
Query: 216 GFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQV-HGLSMKLEACDHVM 274
G +F+ M ++ PN T V V+SAC + G ++ H + +
Sbjct: 215 GLGSEAILLFRSM-LHQGIVPNAATFVGVLSACNHAGLVCEGLRIFHSMQRCYGVTPTIE 391
Query: 275 VVTSLVDMYSKCG 287
T +VD+ + G
Sbjct: 392 HYTCVVDLLGRSG 430
>TC233590 similar to UP|Q6K9T2 (Q6K9T2) Pentatricopeptide repeat-containing
protein-like, partial (5%)
Length = 1094
Score = 156 bits (394), Expect = 3e-38
Identities = 98/290 (33%), Positives = 152/290 (51%), Gaps = 2/290 (0%)
Frame = +2
Query: 311 IAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFSKMQCAGV 370
++G + + E ELF +M DS +W SLI G+A+ G+ EA F +M V
Sbjct: 2 VSGYAVWGDMESGAELFSQMPKS----DSCSWTSLIRGYARNGMGYEALGVFKQMIKHQV 169
Query: 371 APCLKILTSLLSVCGDSCVLRSAKAIHGYALRICVDKDDFLATALVDTYMKCGCVSFARF 430
P L++ L C L+ + IH + + + + + A+V+ Y KCG + AR
Sbjct: 170 RPDQFTLSTCLFACATIASLKHGRQIHAFLVLNNIKPNTIVVCAIVNMYSKCGSLETARR 349
Query: 431 VFDQFDVKPDDPAFWNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHS 490
VF+ F D WN MI G A + Y ML V+PN TFV +L+AC HS
Sbjct: 350 VFN-FIGNKQDVVLWNTMILALAHYGYGIEAIMMLYNMLKIGVKPNKGTFVGILNACCHS 526
Query: 491 GQIERGLRFFR-MIRKYGLDPKPEHFGCVVDLLGRAGQLGEA-RDLVQELAEPPASVFDS 548
G ++ GL+ F+ M ++G+ P EH+ + +LLG+A E+ +DL +P V +S
Sbjct: 527 GLVQEGLQLFKSMTSEHGVVPDQEHYTRLANLLGQARCFNESVKDLQMMDCKPGDHVCNS 706
Query: 549 LLGACRCYLDSNLGEEMAMKLIDIEPKNPAPLVVLSNIYAALGRWSEVER 598
+G CR + + + G E+A LI ++P++ A +LS YAALG+W VE+
Sbjct: 707 SIGVCRMHGNIDHGAEVAAFLIKLQPQSSAAYELLSRTYAALGKWELVEK 856
Score = 91.7 bits (226), Expect = 9e-19
Identities = 57/220 (25%), Positives = 112/220 (50%), Gaps = 3/220 (1%)
Frame = +2
Query: 178 VTAYSKCGVLVSSNKVFENLRVKNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPN 237
V+ Y+ G + S ++F + + ++ + + G +NG VFK M + + +P+
Sbjct: 2 VSGYAVWGDMESGAELFSQMPKSDSCSWTSLIRGYARNGMGYEALGVFKQMIKH-QVRPD 178
Query: 238 KVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFS 297
+ TL + + ACAT+++++ G+Q+H + + +VV ++V+MYSKCG +A VF+
Sbjct: 179 QFTLSTCLFACATIASLKHGRQIHAFLVLNNIKPNTIVVCAIVNMYSKCGSLETARRVFN 358
Query: 298 -RSEKRNLITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCV 356
K++++ WN+MI + A+ + M+ G+ P+ T+ +++ G+
Sbjct: 359 FIGNKQDVVLWNTMILALAHYGYGIEAIMMLYNMLKIGVKPNKGTFVGILNACCHSGLVQ 538
Query: 357 EAFKYFSKMQCA-GVAPCLKILTSLLSVCGDS-CVLRSAK 394
E + F M GV P + T L ++ G + C S K
Sbjct: 539 EGLQLFKSMTSEHGVVPDQEHYTRLANLLGQARCFNESVK 658
Score = 76.3 bits (186), Expect = 4e-14
Identities = 64/264 (24%), Positives = 123/264 (46%), Gaps = 8/264 (3%)
Frame = +2
Query: 92 ELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPNSVTI-VSLLSARDVK 150
ELF +MP+ ++ +++ G +RNG +A+ +F+Q+ +RP+ T+ L + +
Sbjct: 44 ELFSQMPKSDSCSWTSLIRGYARNGMGYEALGVFKQMIKHQVRPDQFTLSTCLFACATIA 223
Query: 151 NQSHVQQVHCLACKLGVEYDVYVSTSLVTAYSKCGVLVSSNKVFENLRVK-NVVTYNAFM 209
+ H +Q+H ++ + V ++V YSKCG L ++ +VF + K +VV +N +
Sbjct: 224 SLKHGRQIHAFLVLNNIKPNTIVVCAIVNMYSKCGSLETARRVFNFIGNKQDVVLWNTMI 403
Query: 210 SGLLQNGFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEA 269
L G+ + +M + + KPNK T V +++AC ++ G Q+ K
Sbjct: 404 LALAHYGYGIEAIMMLYNM-LKIGVKPNKGTFVGILNACCHSGLVQEGLQL----FKSMT 568
Query: 270 CDHVMV-----VTSLVDMYSKCGCWG-SAFDVFSRSEKRNLITWNSMIAGMMMNSESERA 323
+H +V T L ++ + C+ S D+ K NS I M+ +
Sbjct: 569 SEHGVVPDQEHYTRLANLLGQARCFNESVKDLQMMDCKPGDHVCNSSIGVCRMHGNIDHG 748
Query: 324 VELFERMVDEGILPDSATWNSLIS 347
E+ ++ + P S+ L+S
Sbjct: 749 AEVAAFLIK--LQPQSSAAYELLS 814
Score = 41.2 bits (95), Expect = 0.001
Identities = 30/135 (22%), Positives = 61/135 (44%), Gaps = 1/135 (0%)
Frame = +2
Query: 13 NGLYKEALNLYSHLHSSSPTPNTFTFPILLKACSNLSSPSQTQILHAHLFKTGFHSHPHT 72
NG+ EAL ++ + P+ FT L AC+ ++S + +HA L +
Sbjct: 113 NGMGYEALGVFKQMIKHQVRPDQFTLSTCLFACATIASLKHGRQIHAFLVLNNIKPNTIV 292
Query: 73 STALIASYAANTRSFHYALELFDEM-PQPTITAFNAVLSGLSRNGPRGQAVWLFRQIGFW 131
A++ Y + S A +F+ + + + +N ++ L+ G +A+ + +
Sbjct: 293 VCAIVNMY-SKCGSLETARRVFNFIGNKQDVVLWNTMILALAHYGYGIEAIMMLYNMLKI 469
Query: 132 NIRPNSVTIVSLLSA 146
++PN T V +L+A
Sbjct: 470 GVKPNKGTFVGILNA 514
>TC220442 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding protein-like,
partial (25%)
Length = 706
Score = 137 bits (345), Expect = 1e-32
Identities = 69/175 (39%), Positives = 107/175 (60%), Gaps = 2/175 (1%)
Frame = +1
Query: 445 WNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFFR-MI 503
W A+IGG +G A + F +M + PNS TF ++L+ACSH+G E G F M
Sbjct: 19 WTAIIGGLAIHGKGREALDWFTQMQKAGINPNSITFTAILTACSHAGLTEEGKSLFESMS 198
Query: 504 RKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQEL-AEPPASVFDSLLGACRCYLDSNLG 562
Y + P EH+GC+VDL+GRAG L EAR+ ++ + +P A+++ +LL AC+ + LG
Sbjct: 199 SVYNIKPSMEHYGCMVDLMGRAGLLKEAREFIESMPVKPNAAIWGALLNACQLHKHFELG 378
Query: 563 EEMAMKLIDIEPKNPAPLVVLSNIYAALGRWSEVERIRGLITDKGLDKNSGISMI 617
+E+ LI+++P + + L++IYAA G W++V R+R I +GL + G S I
Sbjct: 379 KEIGKILIELDPDHSGRYIHLASIYAAAGEWNQVVRVRSQIKHRGLLNHPGCSSI 543
Score = 43.5 bits (101), Expect = 3e-04
Identities = 44/209 (21%), Positives = 86/209 (41%), Gaps = 5/209 (2%)
Frame = +1
Query: 102 ITAFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPNSVTIVSLLSARDVKNQSHVQQVHCL 161
+ A+ A++ GL+ +G +A+ F Q+ I PNS+T
Sbjct: 10 VCAWTAIIGGLAIHGKGREALDWFTQMQKAGINPNSITF--------------------- 126
Query: 162 ACKLGVEYDVYVSTSLVTAYSKCGVLVSSNKVFENL----RVKNVVTYNAFMSGLL-QNG 216
T+++TA S G+ +FE++ +K + + M L+ + G
Sbjct: 127 -------------TAILTACSHAGLTEEGKSLFESMSSVYNIKPSMEHYGCMVDLMGRAG 267
Query: 217 FHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVV 276
+ + + M + KPN ++++AC + LGK++ + ++L+ DH
Sbjct: 268 LLKEAREFIESMPV----KPNAAIWGALLNACQLHKHFELGKEIGKILIELDP-DHSGRY 432
Query: 277 TSLVDMYSKCGCWGSAFDVFSRSEKRNLI 305
L +Y+ G W V S+ + R L+
Sbjct: 433 IHLASIYAAAGEWNQVVRVRSQIKHRGLL 519
Score = 31.2 bits (69), Expect = 1.4
Identities = 13/38 (34%), Positives = 23/38 (60%)
Frame = +1
Query: 10 LVANGLYKEALNLYSHLHSSSPTPNTFTFPILLKACSN 47
L +G +EAL+ ++ + + PN+ TF +L ACS+
Sbjct: 40 LAIHGKGREALDWFTQMQKAGINPNSITFTAILTACSH 153
>CO979675
Length = 732
Score = 128 bits (321), Expect = 9e-30
Identities = 69/186 (37%), Positives = 108/186 (57%), Gaps = 7/186 (3%)
Frame = -2
Query: 441 DPAFWNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFF 500
D WNAMI G +GD SA ++F EM ++P+ TF++V +ACS+SG GL+
Sbjct: 728 DIVCWNAMISGLAMHGDGASALKMFSEMEKTGIKPDDITFIAVFTACSYSGMAHEGLQLL 549
Query: 501 -RMIRKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQEL------AEPPASVFDSLLGAC 553
+M Y ++PK EH+GC+VDLL RAG GEA +++ + + + L AC
Sbjct: 548 DKMSSLYEIEPKSEHYGCLVDLLSRAGLFGEAMVMIRRITSTSWNGSEETLAWRAFLSAC 369
Query: 554 RCYLDSNLGEEMAMKLIDIEPKNPAPLVVLSNIYAALGRWSEVERIRGLITDKGLDKNSG 613
+ + L E A +L+ +E + V+LSN+YAA G+ S+ R+R ++ +KG+DK G
Sbjct: 368 CNHGQAQLAERAAKRLLRLE-NHSGVYVLLSNLYAASGKHSDARRVRNMMRNKGVDKAPG 192
Query: 614 ISMIEV 619
S +E+
Sbjct: 191 CSSVEI 174
Score = 47.0 bits (110), Expect = 3e-05
Identities = 26/96 (27%), Positives = 49/96 (50%), Gaps = 7/96 (7%)
Frame = -2
Query: 302 RNLITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKY 361
R+++ WN+MI+G+ M+ + A+++F M GI PD T+ ++ + + G+ E +
Sbjct: 731 RDIVCWNAMISGLAMHGDGASALKMFSEMEKTGIKPDDITFIAVFTACSYSGMAHEGLQL 552
Query: 362 FSKMQCA-GVAP------CLKILTSLLSVCGDSCVL 390
KM + P CL L S + G++ V+
Sbjct: 551 LDKMSSLYEIEPKSEHYGCLVDLLSRAGLFGEAMVM 444
Score = 38.9 bits (89), Expect = 0.007
Identities = 30/139 (21%), Positives = 62/139 (44%), Gaps = 7/139 (5%)
Frame = -2
Query: 200 KNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQ 259
+++V +NA +SGL +G +F +M KP+ +T ++V +AC+ G Q
Sbjct: 731 RDIVCWNAMISGLAMHGDGASALKMFSEMEKT-GIKPDDITFIAVFTACSYSGMAHEGLQ 555
Query: 260 -VHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSR------SEKRNLITWNSMIA 312
+ +S E LVD+ S+ G +G A + R + + W + ++
Sbjct: 554 LLDKMSSLYEIEPKSEHYGCLVDLLSRAGLFGEAMVMIRRITSTSWNGSEETLAWRAFLS 375
Query: 313 GMMMNSESERAVELFERMV 331
+ +++ A +R++
Sbjct: 374 ACCNHGQAQLAERAAKRLL 318
Score = 34.7 bits (78), Expect = 0.13
Identities = 14/53 (26%), Positives = 31/53 (58%)
Frame = -2
Query: 102 ITAFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPNSVTIVSLLSARDVKNQSH 154
I +NA++SGL+ +G A+ +F ++ I+P+ +T +++ +A +H
Sbjct: 725 IVCWNAMISGLAMHGDGASALKMFSEMEKTGIKPDDITFIAVFTACSYSGMAH 567
>TC222432
Length = 951
Score = 77.0 bits (188), Expect(2) = 2e-28
Identities = 39/115 (33%), Positives = 66/115 (56%), Gaps = 1/115 (0%)
Frame = +2
Query: 506 YGLDPKPEHFGCVVDLLGRAGQLGEARDLVQELA-EPPASVFDSLLGACRCYLDSNLGEE 564
+G P+ EH+GC+VDLLGRAG L EA +L+Q + + + S L AC + D E
Sbjct: 200 FGFAPQVEHYGCMVDLLGRAGCLDEAENLIQTMPYDANGIILSSFLFACGYFNDVLRAER 379
Query: 565 MAMKLIDIEPKNPAPLVVLSNIYAALGRWSEVERIRGLITDKGLDKNSGISMIEV 619
+ +++ ++ V+L N+YA RW++VE ++ ++ +G K S+IE+
Sbjct: 380 VLKEVVKMDEDVAGNYVMLRNLYATRQRWTDVEDVKQMMKKRGTSKEVACSVIEI 544
Score = 67.4 bits (163), Expect(2) = 2e-28
Identities = 29/66 (43%), Positives = 41/66 (61%)
Frame = +3
Query: 441 DPAFWNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFF 500
+ A WNA+I G+ NG + A EVF M++E PN T + VLSAC+H G +E G R+F
Sbjct: 3 ETASWNALINGFAVNGCAKEALEVFARMIEEGFGPNEVTMIGVLSACNHCGLVEEGRRWF 182
Query: 501 RMIRKY 506
+ +Y
Sbjct: 183 NAMERY 200
Score = 44.3 bits (103), Expect = 2e-04
Identities = 19/61 (31%), Positives = 37/61 (60%)
Frame = +3
Query: 306 TWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFSKM 365
+WN++I G +N ++ A+E+F RM++EG P+ T ++S G+ E ++F+ M
Sbjct: 12 SWNALINGFAVNGCAKEALEVFARMIEEGFGPNEVTMIGVLSACNHCGLVEEGRRWFNAM 191
Query: 366 Q 366
+
Sbjct: 192 E 194
Score = 38.9 bits (89), Expect = 0.007
Identities = 18/47 (38%), Positives = 28/47 (59%)
Frame = +3
Query: 338 DSATWNSLISGFAQKGVCVEAFKYFSKMQCAGVAPCLKILTSLLSVC 384
++A+WN+LI+GFA G EA + F++M G P + +LS C
Sbjct: 3 ETASWNALINGFAVNGCAKEALEVFARMIEEGFGPNEVTMIGVLSAC 143
Score = 33.1 bits (74), Expect = 0.38
Identities = 19/58 (32%), Positives = 33/58 (56%), Gaps = 2/58 (3%)
Frame = +3
Query: 204 TYNAFMSGLLQNGFHRVVFDVFKDMTMNLEEK--PNKVTLVSVVSACATLSNIRLGKQ 259
++NA ++G NG + +VF M +EE PN+VT++ V+SAC + G++
Sbjct: 12 SWNALINGFAVNGCAKEALEVFARM---IEEGFGPNEVTMIGVLSACNHCGLVEEGRR 176
Score = 29.6 bits (65), Expect = 4.2
Identities = 14/43 (32%), Positives = 28/43 (64%)
Frame = +3
Query: 104 AFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPNSVTIVSLLSA 146
++NA+++G + NG +A+ +F ++ PN VT++ +LSA
Sbjct: 12 SWNALINGFAVNGCAKEALEVFARMIEEGFGPNEVTMIGVLSA 140
>TC232145
Length = 817
Score = 120 bits (301), Expect = 2e-27
Identities = 65/151 (43%), Positives = 94/151 (62%), Gaps = 2/151 (1%)
Frame = +3
Query: 471 EMVQPNSATFVSVLSACSHSGQIERGLRFFR-MIRKYGLDPKPEHFGCVVDLLGRAGQLG 529
E V P TFVSVLSACSH+G+I+ G ++F M + + P EH+ C+VDLLGR G+L
Sbjct: 3 EGVVPEYITFVSVLSACSHTGKIDDGFKYFNSMANVHNIKPGLEHYACMVDLLGRVGRLE 182
Query: 530 EARDLVQELA-EPPASVFDSLLGACRCYLDSNLGEEMAMKLIDIEPKNPAPLVVLSNIYA 588
EA ++ + EP + V+ +LLGAC + + +G E+A +L +EP NP ++LSNIY
Sbjct: 183 EACRFIESMPFEPDSLVWGALLGACGKHANVEMGREVAERLFKLEPDNPGNYMLLSNIYI 362
Query: 589 ALGRWSEVERIRGLITDKGLDKNSGISMIEV 619
G E + +R L+ G+ K SG S I+V
Sbjct: 363 RHGMLEEADEVRRLMGINGVRKESGCSWIDV 455
Score = 37.0 bits (84), Expect = 0.026
Identities = 34/134 (25%), Positives = 61/134 (45%), Gaps = 2/134 (1%)
Frame = +3
Query: 333 EGILPDSATWNSLISGFAQKGVCVEAFKYFSKM-QCAGVAPCLKILTSLLSVCGDSCVLR 391
EG++P+ T+ S++S + G + FKYF+ M + P L+ ++ + G L
Sbjct: 3 EGVVPEYITFVSVLSACSHTGKIDDGFKYFNSMANVHNIKPGLEHYACMVDLLGRVGRLE 182
Query: 392 SAKAIHGYALRICVDKDDFLATALVDTYMKCGCVSFARFVFDQ-FDVKPDDPAFWNAMIG 450
A + + + D + AL+ K V R V ++ F ++PD+P + +
Sbjct: 183 EACR---FIESMPFEPDSLVWGALLGACGKHANVEMGREVAERLFKLEPDNPGNYMLLSN 353
Query: 451 GYGTNGDYESAFEV 464
Y +G E A EV
Sbjct: 354 IYIRHGMLEEADEV 395
>AW396441
Length = 459
Score = 119 bits (299), Expect = 3e-27
Identities = 57/152 (37%), Positives = 90/152 (58%)
Frame = +3
Query: 379 SLLSVCGDSCVLRSAKAIHGYALRICVDKDDFLATALVDTYMKCGCVSFARFVFDQFDVK 438
++++ + LR + H +++ +D D F+ +LVD Y KCG + + F + +
Sbjct: 9 AVIAAASNIASLRHGQQFHNQVIKMGLDDDPFVTNSLVDMYAKCGSIEESHKAFSSTNQR 188
Query: 439 PDDPAFWNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGLR 498
D A WN+MI Y +GD A EVF M+ E V+PN TFV +LSACSH+G ++ G
Sbjct: 189 --DIACWNSMISTYAQHGDAAKALEVFERMIMEGVKPNYVTFVGLLSACSHAGLLDLGFH 362
Query: 499 FFRMIRKYGLDPKPEHFGCVVDLLGRAGQLGE 530
F + K+G++P +H+ C+V LLGRAG++ E
Sbjct: 363 HFESMSKFGIEPXIDHYACMVSLLGRAGKIYE 458
Score = 95.1 bits (235), Expect = 8e-20
Identities = 46/143 (32%), Positives = 83/143 (57%)
Frame = +3
Query: 243 SVVSACATLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSEKR 302
+V++A + ++++R G+Q H +K+ D V SLVDMY+KCG + FS + +R
Sbjct: 9 AVIAAASNIASLRHGQQFHNQVIKMGLDDDPFVTNSLVDMYAKCGSIEESHKAFSSTNQR 188
Query: 303 NLITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYF 362
++ WNSMI+ + ++ +A+E+FERM+ EG+ P+ T+ L+S + G+ F +F
Sbjct: 189 DIACWNSMISTYAQHGDAAKALEVFERMIMEGVKPNYVTFVGLLSACSHAGLLDLGFHHF 368
Query: 363 SKMQCAGVAPCLKILTSLLSVCG 385
M G+ P + ++S+ G
Sbjct: 369 ESMSKFGIEPXIDHYACMVSLLG 437
Score = 71.2 bits (173), Expect = 1e-12
Identities = 38/115 (33%), Positives = 65/115 (56%)
Frame = +3
Query: 143 LLSARDVKNQSHVQQVHCLACKLGVEYDVYVSTSLVTAYSKCGVLVSSNKVFENLRVKNV 202
+ +A ++ + H QQ H K+G++ D +V+ SLV Y+KCG + S+K F + +++
Sbjct: 15 IAAASNIASLRHGQQFHNQVIKMGLDDDPFVTNSLVDMYAKCGSIEESHKAFSSTNQRDI 194
Query: 203 VTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLG 257
+N+ +S Q+G +VF+ M M KPN VT V ++SAC+ + LG
Sbjct: 195 ACWNSMISTYAQHGDAAKALEVFERMIME-GVKPNYVTFVGLLSACSHAGLLDLG 356
Score = 52.4 bits (124), Expect = 6e-07
Identities = 31/109 (28%), Positives = 55/109 (50%)
Frame = +3
Query: 38 FPILLKACSNLSSPSQTQILHAHLFKTGFHSHPHTSTALIASYAANTRSFHYALELFDEM 97
F ++ A SN++S Q H + K G P + +L+ YA S + + F
Sbjct: 3 FAAVIAAASNIASLRHGQQFHNQVIKMGLDDDPFVTNSLVDMYA-KCGSIEESHKAFSST 179
Query: 98 PQPTITAFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPNSVTIVSLLSA 146
Q I +N+++S +++G +A+ +F ++ ++PN VT V LLSA
Sbjct: 180 NQRDIACWNSMISTYAQHGDAAKALEVFERMIMEGVKPNYVTFVGLLSA 326
>TC224522 weakly similar to UP|Q9LUL5 (Q9LUL5) Emb|CAB66100.1, partial (18%)
Length = 534
Score = 117 bits (293), Expect = 2e-26
Identities = 54/143 (37%), Positives = 95/143 (65%), Gaps = 2/143 (1%)
Frame = +2
Query: 479 TFVSVLSACSHSGQIERGLRFFR-MIRKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQE 537
TF+S+LSAC +G++ + F M+ YG+ P+ EH+ C+VD++ RAGQL A ++ E
Sbjct: 11 TFLSLLSACCRAGKVNESMNLFSLMVDNYGIPPRSEHYACLVDVMSRAGQLQRACKIINE 190
Query: 538 LA-EPPASVFDSLLGACRCYLDSNLGEEMAMKLIDIEPKNPAPLVVLSNIYAALGRWSEV 596
+ + +S++ ++L AC +L+ LGE A ++++++P N V+LSNIYAA G+W +V
Sbjct: 191 MPFKADSSIWGAVLAACSVHLNVELGELAARRILNLDPFNSGAYVMLSNIYAAAGKWKDV 370
Query: 597 ERIRGLITDKGLDKNSGISMIEV 619
RIR L+ ++G+ K + S +++
Sbjct: 371 HRIRVLMKEQGVKKQTAYSWLQI 439
>BG790334 weakly similar to GP|2160154|gb| F19K23.18 gene product
{Arabidopsis thaliana}, partial (19%)
Length = 390
Score = 114 bits (285), Expect = 1e-25
Identities = 58/123 (47%), Positives = 84/123 (68%), Gaps = 2/123 (1%)
Frame = +2
Query: 473 VQPNSATFVSVLSACSHSGQIERGLRFFR-MIRKYGLDPKPEHFGCVVDLLGRAGQLGEA 531
+ P TF+SVL+AC+H+G +E G R F+ MI YG++P+ EHF +VD+LGR GQL EA
Sbjct: 20 IHPTYITFISVLNACAHAGLVEEGWRQFKSMINDYGIEPRVEHFASLVDILGRQGQLQEA 199
Query: 532 RDLVQELA-EPPASVFDSLLGACRCYLDSNLGEEMAMKLIDIEPKNPAPLVVLSNIYAAL 590
DL+ + +P +V+ +LLGACR + + L A LI +EP++ AP V+L N+YA L
Sbjct: 200 MDLINTMPFKPDKAVWGALLGACRVHNNVELALVAADALIRLEPESSAPYVLLYNMYANL 379
Query: 591 GRW 593
G+W
Sbjct: 380 GQW 388
>TC234676
Length = 914
Score = 114 bits (284), Expect = 2e-25
Identities = 79/280 (28%), Positives = 124/280 (44%)
Frame = +1
Query: 257 GKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSEKRNLITWNSMIAGMMM 316
G Q+H ++K +V V +SL+ MYSKCG + +F E+RN+I+W +M
Sbjct: 13 GMQIHAYALKHWFLPNVSVASSLMTMYSKCGVVEYSRRLFDNMEQRNVISWTAM------ 174
Query: 317 NSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFSKMQCAGVAPCLKI 376
I + + G EA MQ + P
Sbjct: 175 -----------------------------IDSYIENGYLCEALGVIRSMQLSKHRPDSVA 267
Query: 377 LTSLLSVCGDSCVLRSAKAIHGYALRICVDKDDFLATALVDTYMKCGCVSFARFVFDQFD 436
+ +LSVCG+ +++ K IHG L+ F++ L++ Y G ++ A VF+
Sbjct: 268 IGRMLSVCGERKLVKLGKEIHGQILKRDFTSVHFVSAELINMYGFFGDINKANLVFNAVP 447
Query: 437 VKPDDPAFWNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERG 496
VK W A+I YG N Y+ A +F +M PN TF ++LS C +G ++
Sbjct: 448 VK--GSMTWTALIRAYGYNELYQDAVNLFDQM---RYSPNHFTFEAILSICDKAGFVDDA 612
Query: 497 LRFFRMIRKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQ 536
R F + +Y ++ EHF +V LL GQL +A+ Q
Sbjct: 613 CRIFNSMPRYKIEASKEHFAIMVRLLTHNGQLEKAQRFEQ 732
Score = 100 bits (249), Expect = 2e-21
Identities = 59/209 (28%), Positives = 111/209 (52%)
Frame = +1
Query: 157 QVHCLACKLGVEYDVYVSTSLVTAYSKCGVLVSSNKVFENLRVKNVVTYNAFMSGLLQNG 216
Q+H A K +V V++SL+T YSKCGV+ S ++F+N+ +NV+++ A + ++NG
Sbjct: 19 QIHAYALKHWFLPNVSVASSLMTMYSKCGVVEYSRRLFDNMEQRNVISWTAMIDSYIENG 198
Query: 217 FHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVV 276
+ V + M ++ + +P+ V + ++S C ++LGK++HG +K + V
Sbjct: 199 YLCEALGVIRSMQLS-KHRPDSVAIGRMLSVCGERKLVKLGKEIHGQILKRDFTSVHFVS 375
Query: 277 TSLVDMYSKCGCWGSAFDVFSRSEKRNLITWNSMIAGMMMNSESERAVELFERMVDEGIL 336
L++MY G A VF+ + +TW ++I N + AV LF++M
Sbjct: 376 AELINMYGFFGDINKANLVFNAVPVKGSMTWTALIRAYGYNELYQDAVNLFDQM---RYS 546
Query: 337 PDSATWNSLISGFAQKGVCVEAFKYFSKM 365
P+ T+ +++S + G +A + F+ M
Sbjct: 547 PNHFTFEAILSICDKAGFVDDACRIFNSM 633
Score = 31.6 bits (70), Expect = 1.1
Identities = 18/49 (36%), Positives = 27/49 (54%), Gaps = 4/49 (8%)
Frame = +1
Query: 1 MKSDITVTKLVA----NGLYKEALNLYSHLHSSSPTPNTFTFPILLKAC 45
+K +T T L+ N LY++A+NL+ + S PN FTF +L C
Sbjct: 448 VKGSMTWTALIRAYGYNELYQDAVNLFDQMRYS---PNHFTFEAILSIC 585
>CD397389 similar to GP|6691205|gb| F1K23.11 {Arabidopsis thaliana}, partial
(16%)
Length = 626
Score = 114 bits (284), Expect = 2e-25
Identities = 63/177 (35%), Positives = 101/177 (56%), Gaps = 4/177 (2%)
Frame = -2
Query: 445 WNAMIGGYGTNGDYESAFEVFYEMLDEM-VQPNSATFVSVLSACSHSGQIERGLRFFR-M 502
W +M+ GYG NG + A E+F +M E + PN T +S LSAC+H+G +++G + M
Sbjct: 625 WTSMMDGYGKNGFPDEALELFVKMPTEYGIVPNYVTLLSALSACAHAGLVDKGWEIIQSM 446
Query: 503 IRKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQELAEPPAS-VFDSLLGACRCYLDSNL 561
+Y + P EH+ C+VDLLGRAG L +A + + + E P S V+ +LL +CR + + L
Sbjct: 445 ENEYLVKPGMEHYACMVDLLGRAGMLNQAWEFIMRIPEKPISDVWAALLSSCRLHGNIEL 266
Query: 562 GEEMAMKLIDIEPK-NPAPLVVLSNIYAALGRWSEVERIRGLITDKGLDKNSGISMI 617
+ A +L + P V LSN A G W V +R ++ ++G+ K++ S +
Sbjct: 265 AKLAANELFKLNATGRPGAYVALSNTLVAAG*WESVTELREIMKERGISKDTXRSWV 95
Score = 37.0 bits (84), Expect = 0.026
Identities = 28/121 (23%), Positives = 52/121 (42%), Gaps = 2/121 (1%)
Frame = -2
Query: 205 YNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQ-VHGL 263
+ + M G +NGF ++F M PN VTL+S +SACA + G + + +
Sbjct: 625 WTSMMDGYGKNGFPDEALELFVKMPTEYGIVPNYVTLLSALSACAHAGLVDKGWEIIQSM 446
Query: 264 SMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSR-SEKRNLITWNSMIAGMMMNSESER 322
+ + +VD+ + G A++ R EK W ++++ ++ E
Sbjct: 445 ENEYLVKPGMEHYACMVDLLGRAGMLNQAWEFIMRIPEKPISDVWAALLSSCRLHGNIEL 266
Query: 323 A 323
A
Sbjct: 265 A 263
Score = 34.7 bits (78), Expect = 0.13
Identities = 20/61 (32%), Positives = 32/61 (51%), Gaps = 1/61 (1%)
Frame = -2
Query: 307 WNSMIAGMMMNSESERAVELFERMVDE-GILPDSATWNSLISGFAQKGVCVEAFKYFSKM 365
W SM+ G N + A+ELF +M E GI+P+ T S +S A G+ + ++ M
Sbjct: 625 WTSMMDGYGKNGFPDEALELFVKMPTEYGIVPNYVTLLSALSACAHAGLVDKGWEIIQSM 446
Query: 366 Q 366
+
Sbjct: 445 E 443
>TC212581 weakly similar to UP|Q6K957 (Q6K957) Pentatricopeptide (PPR)
repeat-containing protein-like, partial (8%)
Length = 1044
Score = 109 bits (273), Expect = 3e-24
Identities = 89/283 (31%), Positives = 141/283 (49%), Gaps = 3/283 (1%)
Frame = +3
Query: 38 FPILLKACSNLSSPSQTQILHAHLFKTGFHSHPHTSTALIASYAANTRSFHYALELFDEM 97
F + L SNL + +HA L + S H + L++ YA F +A LFD+M
Sbjct: 114 FLLSLAKTSNLVTLRHCNQIHAKLIASQCISQTHLTNNLLSLYA-KFGHFRHAHLLFDQM 290
Query: 98 PQPTITAFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPNSVTIVSLLSARDVKNQSHVQ- 156
PQ + + ++S R G +A +F I N RPN T LL A + +V
Sbjct: 291 PQRNVFTWTTLISSHFRTGSLPKAFEMFNHICALNERPNEYTFSVLLRACATPSLWNVGL 470
Query: 157 QVHCLACKLGVEYDVYVSTSLVTAYSKCGV-LVSSNKVFENLRVKNVVTYNAFMSGLLQN 215
Q+H L + G+E + + +S+V Y G L + F +L +++V +N +SG +
Sbjct: 471 QIHGLLVRSGLERNKFSGSSIVYMYFNSGSNLGDACCAFHDLLERDLVAWNVMISGFARV 650
Query: 216 GFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMV 275
G +V +F +M KP+ T VS++ C++L + KQ+HGL+ K A V+V
Sbjct: 651 GDFSMVHRLFSEMWGVEGLKPDDCTFVSLLKCCSSLKEL---KQIHGLASKFGAEVDVVV 821
Query: 276 VTSLVDMYSKCGCWGSAFDVF-SRSEKRNLITWNSMIAGMMMN 317
+LVD+ +K G S VF S+ EK N + W+ +I+G MN
Sbjct: 822 GNALVDL*NKHGDVSSCRKVFDSKKEKYNFV-WSLIISGYSMN 947
Score = 87.4 bits (215), Expect = 2e-17
Identities = 55/203 (27%), Positives = 104/203 (51%), Gaps = 1/203 (0%)
Frame = +3
Query: 14 GLYKEALNLYSHLHSSSPTPNTFTFPILLKACSNLSSPSQTQILHAHLFKTGFHSHPHTS 73
G +A +++H+ + + PN +TF +LL+AC+ S + +H L ++G + +
Sbjct: 345 GSLPKAFEMFNHICALNERPNEYTFSVLLRACATPSLWNVGLQIHGLLVRSGLERNKFSG 524
Query: 74 TALIASYAANTRSFHYALELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWLFRQI-GFWN 132
++++ Y + + A F ++ + + A+N ++SG +R G LF ++ G
Sbjct: 525 SSIVYMYFNSGSNLGDACCAFHDLLERDLVAWNVMISGFARVGDFSMVHRLFSEMWGVEG 704
Query: 133 IRPNSVTIVSLLSARDVKNQSHVQQVHCLACKLGVEYDVYVSTSLVTAYSKCGVLVSSNK 192
++P+ T VSLL + ++Q+H LA K G E DV V +LV +K G + S K
Sbjct: 705 LKPDDCTFVSLLKC--CSSLKELKQIHGLASKFGAEVDVVVGNALVDL*NKHGDVSSCRK 878
Query: 193 VFENLRVKNVVTYNAFMSGLLQN 215
VF++ + K ++ +SG N
Sbjct: 879 VFDSKKEKYNFVWSLIISGYSMN 947
Score = 72.0 bits (175), Expect = 7e-13
Identities = 64/299 (21%), Positives = 115/299 (38%), Gaps = 7/299 (2%)
Frame = +3
Query: 249 ATLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSEKRNLITWN 308
+ L +R Q+H + + + +L+ +Y+K G + A +F + +RN+ T
Sbjct: 138 SNLVTLRHCNQIHAKLIASQCISQTHLTNNLLSLYAKFGHFRHAHLLFDQMPQRNVFT-- 311
Query: 309 SMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFSKMQCA 368
W +LIS + G +AF+ F+ +
Sbjct: 312 ---------------------------------WTTLISSHFRTGSLPKAFEMFNHICAL 392
Query: 369 GVAPCLKILTSLLSVCGDSCVLRSAKAIHGYALRICVDKDDFLATALVDTYMKCG----- 423
P + LL C + IHG +R ++++ F +++V Y G
Sbjct: 393 NERPNEYTFSVLLRACATPSLWNVGLQIHGLLVRSGLERNKFSGSSIVYMYFNSGSNLGD 572
Query: 424 -CVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNGDYESAFEVFYEMLD-EMVQPNSATFV 481
C +F D+ D WN MI G+ GD+ +F EM E ++P+ TFV
Sbjct: 573 ACCAF-------HDLLERDLVAWNVMISGFARVGDFSMVHRLFSEMWGVEGLKPDDCTFV 731
Query: 482 SVLSACSHSGQIERGLRFFRMIRKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQELAE 540
S+L CS +++ + + K+G + +VDL + G + R + E
Sbjct: 732 SLLKCCSSLKELK---QIHGLASKFGAEVDVVVGNALVDL*NKHGDVSSCRKVFDSKKE 899
>BE802290
Length = 481
Score = 105 bits (263), Expect = 5e-23
Identities = 57/155 (36%), Positives = 90/155 (57%), Gaps = 2/155 (1%)
Frame = -1
Query: 465 FYEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFFR-MIRKYGLDPKPEHFGCVVDLLG 523
F + + +PNS F+++ ACSH+ E G F M Y + P EH+GC+VDL+G
Sbjct: 481 FTQCRKQKSKPNSIPFMAISPACSHAALTEEGTSSFESMSSVYNIKPSMEHYGCMVDLMG 302
Query: 524 RAGQLGEARDLVQEL-AEPPASVFDSLLGACRCYLDSNLGEEMAMKLIDIEPKNPAPLVV 582
AG L EAR+ V+ + +P A+++ +LL AC+ + LG+E I+++P + +
Sbjct: 301 GAGLLKEAREFVESMPVKPNAAIWGALLNACQLHKYFELGKETGKIQIELDPDHSGRYIH 122
Query: 583 LSNIYAALGRWSEVERIRGLITDKGLDKNSGISMI 617
L++IYAA W++V R+R I +GL SG S I
Sbjct: 121 LASIYAAAREWNQVTRVRSQIKHRGLLNYSGCSSI 17
Score = 30.4 bits (67), Expect = 2.4
Identities = 18/83 (21%), Positives = 38/83 (45%)
Frame = -1
Query: 226 KDMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSK 285
++ ++ KPN ++++AC LGK+ + ++L+ DH L +Y+
Sbjct: 277 REFVESMPVKPNAAIWGALLNACQLHKYFELGKETGKIQIELDP-DHSGRYIHLASIYAA 101
Query: 286 CGCWGSAFDVFSRSEKRNLITWN 308
W V S+ + R L+ ++
Sbjct: 100 AREWNQVTRVRSQIKHRGLLNYS 32
>CK769031
Length = 827
Score = 76.3 bits (186), Expect(2) = 1e-22
Identities = 40/101 (39%), Positives = 62/101 (60%), Gaps = 7/101 (6%)
Frame = +2
Query: 450 GGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFFRMIRK--YG 507
GG NG + A + F ++++ +PN TF++V +AC H+G ++ G ++F + Y
Sbjct: 11 GGLAINGYGKEALKRFEDLVESGARPNEVTFLAVYTACCHNGLVDEGRKYFNEMTSPHYN 190
Query: 508 LDPKPEHFGCVVDLLGRAGQLGEARDLV-----QELAEPPA 543
L P EH+GC+VDLL RAG +GEA +L+ QEL+ PA
Sbjct: 191 LSPCLEHYGCMVDLLCRAGLVGEAVELIKIFRSQELS*VPA 313
Score = 48.9 bits (115), Expect(2) = 1e-22
Identities = 27/72 (37%), Positives = 41/72 (56%)
Frame = +1
Query: 548 SLLGACRCYLDSNLGEEMAMKLIDIEPKNPAPLVVLSNIYAALGRWSEVERIRGLITDKG 607
+LL AC + +EM L ++E ++ V+LSN+YA +W+EV IR L+ KG
Sbjct: 295 ALLSACNT*GNVGFTQEMLKSLQNVEFQDSGIYVLLSNLYATNKKWAEVRSIRRLMKQKG 474
Query: 608 LDKNSGISMIEV 619
+ K G S+I V
Sbjct: 475 ISKAPGSSIISV 510
>TC234029 similar to UP|Q9LN01 (Q9LN01) T6D22.15, partial (32%)
Length = 732
Score = 103 bits (258), Expect = 2e-22
Identities = 49/114 (42%), Positives = 75/114 (64%), Gaps = 1/114 (0%)
Frame = +2
Query: 507 GLDPKPEHFGCVVDLLGRAGQLGEARDLVQELA-EPPASVFDSLLGACRCYLDSNLGEEM 565
G+ PK +H+GC++DLL R+G+ EA+ L+ + EP +++ SLL ACR + GE +
Sbjct: 2 GISPKLQHYGCMIDLLARSGKFDEAKVLMGNMEMEPDGAIWGSLLNACRIHGQVEFGEYV 181
Query: 566 AMKLIDIEPKNPAPLVVLSNIYAALGRWSEVERIRGLITDKGLDKNSGISMIEV 619
A +L ++EP+N V+LSNIYA GRW +V +IR + DKG+ K G + IE+
Sbjct: 182 AERLFELEPENSGAYVLLSNIYAGAGRWDDVAKIRTKLNDKGMKKVPGCTSIEI 343
>BI893521
Length = 423
Score = 103 bits (256), Expect = 3e-22
Identities = 51/128 (39%), Positives = 81/128 (62%), Gaps = 2/128 (1%)
Frame = +1
Query: 445 WNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFFR-MI 503
W ++I G+ +G A E+FYEML+ V+PN T+++VLSACSH G I+ + F M
Sbjct: 40 WTSIISGFAKHGFATKALELFYEMLEIGVKPNEVTYIAVLSACSHVGLIDEAWKHFNSMH 219
Query: 504 RKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQELA-EPPASVFDSLLGACRCYLDSNLG 562
+ + P+ EH+ C+VDLLGR+G L EA + + + + A V+ + LG+CR + ++ LG
Sbjct: 220 YNHSISPRMEHYACMVDLLGRSGLLLEAIEFINSMPFDADALVWRTFLGSCRVHRNTKLG 399
Query: 563 EEMAMKLI 570
E A K++
Sbjct: 400 EHAAKKIL 423
Score = 56.2 bits (134), Expect = 4e-08
Identities = 30/100 (30%), Positives = 59/100 (59%), Gaps = 1/100 (1%)
Frame = +1
Query: 295 VFSRSEKRNLITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGV 354
VF+ RN+ITW S+I+G + + +A+ELF M++ G+ P+ T+ +++S + G+
Sbjct: 4 VFNDMGYRNVITWTSIISGFAKHGFATKALELFYEMLEIGVKPNEVTYIAVLSACSHVGL 183
Query: 355 CVEAFKYFSKMQC-AGVAPCLKILTSLLSVCGDSCVLRSA 393
EA+K+F+ M ++P ++ ++ + G S +L A
Sbjct: 184 IDEAWKHFNSMHYNHSISPRMEHYACMVDLLGRSGLLLEA 303
Score = 46.6 bits (109), Expect = 3e-05
Identities = 20/58 (34%), Positives = 40/58 (68%)
Frame = +1
Query: 192 KVFENLRVKNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSACA 249
+VF ++ +NV+T+ + +SG ++GF ++F +M + + KPN+VT ++V+SAC+
Sbjct: 1 QVFNDMGYRNVITWTSIISGFAKHGFATKALELFYEM-LEIGVKPNEVTYIAVLSACS 171
Score = 35.8 bits (81), Expect = 0.058
Identities = 15/55 (27%), Positives = 36/55 (65%)
Frame = +1
Query: 92 ELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPNSVTIVSLLSA 146
++F++M + + +++SG +++G +A+ LF ++ ++PN VT +++LSA
Sbjct: 1 QVFNDMGYRNVITWTSIISGFAKHGFATKALELFYEMLEIGVKPNEVTYIAVLSA 165
>AW317686
Length = 447
Score = 102 bits (255), Expect = 4e-22
Identities = 53/146 (36%), Positives = 89/146 (60%), Gaps = 5/146 (3%)
Frame = +1
Query: 435 FDVKP-DDPAFWNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQI 493
F++ P D WN +I G+ NG E + VF M++ V+PN TF+ VLS C+H+G
Sbjct: 10 FEMAPMRDVVTWNTLITGFAQNGHGEESLAVFRRMIEAKVEPNHVTFLGVLSGCNHAGLD 189
Query: 494 ERGLRFFRMI-RKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQELAE---PPASVFDSL 549
GL+ ++ R+YG+ PK EH+ ++DLLGR +L EA L++++ + +V+ ++
Sbjct: 190 NEGLQLVDLMERQYGVKPKAEHYALLIDLLGRRNRLMEAMSLIEKVPDGIKNHIAVWGAV 369
Query: 550 LGACRCYLDSNLGEEMAMKLIDIEPK 575
LGAC + + +L + A KL ++EP+
Sbjct: 370 LGACXVHGNLDLARKAAEKLFELEPE 447
Score = 47.8 bits (112), Expect = 1e-05
Identities = 24/93 (25%), Positives = 48/93 (50%), Gaps = 1/93 (1%)
Frame = +1
Query: 294 DVFSRSEKRNLITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKG 353
++F + R+++TWN++I G N E ++ +F RM++ + P+ T+ ++SG G
Sbjct: 4 NLFEMAPMRDVVTWNTLITGFAQNGHGEESLAVFRRMIEAKVEPNHVTFLGVLSGCNHAG 183
Query: 354 VCVEAFKYFSKMQCA-GVAPCLKILTSLLSVCG 385
+ E + M+ GV P + L+ + G
Sbjct: 184 LDNEGLQLVDLMERQYGVKPKAEHYALLIDLLG 282
Score = 42.7 bits (99), Expect = 5e-04
Identities = 34/147 (23%), Positives = 70/147 (47%), Gaps = 7/147 (4%)
Frame = +1
Query: 193 VFENLRVKNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSAC--AT 250
+FE +++VVT+N ++G QNG VF+ M + + +PN VT + V+S C A
Sbjct: 7 LFEMAPMRDVVTWNTLITGFAQNGHGEESLAVFRRM-IEAKVEPNHVTFLGVLSGCNHAG 183
Query: 251 LSN-----IRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSEKRNLI 305
L N + L ++ +G+ K E H ++ L+ ++ S + K ++
Sbjct: 184 LDNEGLQLVDLMERQYGVKPKAE---HYALLIDLLGRRNRLMEAMSLIEKVPDGIKNHIA 354
Query: 306 TWNSMIAGMMMNSESERAVELFERMVD 332
W +++ ++ + A + E++ +
Sbjct: 355 VWGAVLGACXVHGNLDLARKAAEKLFE 435
Score = 37.0 bits (84), Expect = 0.026
Identities = 21/57 (36%), Positives = 29/57 (50%)
Frame = +1
Query: 328 ERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFSKMQCAGVAPCLKILTSLLSVC 384
E + + + D TWN+LI+GFAQ G E+ F +M A V P +LS C
Sbjct: 1 ENLFEMAPMRDVVTWNTLITGFAQNGHGEESLAVFRRMIEAKVEPNHVTFLGVLSGC 171
Score = 35.4 bits (80), Expect = 0.076
Identities = 15/53 (28%), Positives = 32/53 (60%)
Frame = +1
Query: 93 LFDEMPQPTITAFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPNSVTIVSLLS 145
LF+ P + +N +++G ++NG +++ +FR++ + PN VT + +LS
Sbjct: 7 LFEMAPMRDVVTWNTLITGFAQNGHGEESLAVFRRMIEAKVEPNHVTFLGVLS 165
>AW234248
Length = 399
Score = 102 bits (253), Expect = 7e-22
Identities = 53/127 (41%), Positives = 76/127 (59%), Gaps = 1/127 (0%)
Frame = +2
Query: 413 TALVDTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNGDYESAFEVFYEMLDEM 472
T+LVD Y KCG + AR +F + + A WNAMI G +G+ E A + F EM
Sbjct: 8 TSLVDMYAKCGNIEDARGLFKRTNTSRI--ASWNAMIVGLAQHGNAEEALQFFEEMKSRG 181
Query: 473 VQPNSATFVSVLSACSHSGQIERGL-RFFRMIRKYGLDPKPEHFGCVVDLLGRAGQLGEA 531
V P+ TF+ VLSACSHSG + F+ M + YG++P+ EH+ C+VD L RAG++ EA
Sbjct: 182 VTPDRVTFIGVLSACSHSGLVSEAYENFYSMQKIYGIEPEIEHYSCLVDALSRAGRIREA 361
Query: 532 RDLVQEL 538
++ +
Sbjct: 362 EKVISSM 382
Score = 78.6 bits (192), Expect = 8e-15
Identities = 40/123 (32%), Positives = 69/123 (55%), Gaps = 1/123 (0%)
Frame = +2
Query: 275 VVTSLVDMYSKCGCWGSAFDVFSRSEKRNLITWNSMIAGMMMNSESERAVELFERMVDEG 334
V+TSLVDMY+KCG A +F R+ + +WN+MI G+ + +E A++ FE M G
Sbjct: 2 VMTSLVDMYAKCGNIEDARGLFKRTNTSRIASWNAMIVGLAQHGNAEEALQFFEEMKSRG 181
Query: 335 ILPDSATWNSLISGFAQKGVCVEAFKYFSKMQ-CAGVAPCLKILTSLLSVCGDSCVLRSA 393
+ PD T+ ++S + G+ EA++ F MQ G+ P ++ + L+ + +R A
Sbjct: 182 VTPDRVTFIGVLSACSHSGLVSEAYENFYSMQKIYGIEPEIEHYSCLVDALSRAGRIREA 361
Query: 394 KAI 396
+ +
Sbjct: 362 EKV 370
Score = 51.6 bits (122), Expect = 1e-06
Identities = 33/122 (27%), Positives = 60/122 (49%), Gaps = 7/122 (5%)
Frame = +2
Query: 173 VSTSLVTAYSKCGVLVSSNKVFENLRVKNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNL 232
V TSLV Y+KCG + + +F+ + ++NA + GL Q+G F++M +
Sbjct: 2 VMTSLVDMYAKCGNIEDARGLFKRTNTSRIASWNAMIVGLAQHGNAEEALQFFEEM-KSR 178
Query: 233 EEKPNKVTLVSVVSACA-------TLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSK 285
P++VT + V+SAC+ N ++++G+ ++E + LVD S+
Sbjct: 179 GVTPDRVTFIGVLSACSHSGLVSEAYENFYSMQKIYGIEPEIEH------YSCLVDALSR 340
Query: 286 CG 287
G
Sbjct: 341 AG 346
Score = 28.9 bits (63), Expect = 7.1
Identities = 14/41 (34%), Positives = 22/41 (53%)
Frame = +2
Query: 7 VTKLVANGLYKEALNLYSHLHSSSPTPNTFTFPILLKACSN 47
+ L +G +EAL + + S TP+ TF +L ACS+
Sbjct: 110 IVGLAQHGNAEEALQFFEEMKSRGVTPDRVTFIGVLSACSH 232
>BE211268
Length = 422
Score = 100 bits (250), Expect = 1e-21
Identities = 52/135 (38%), Positives = 81/135 (59%), Gaps = 2/135 (1%)
Frame = +1
Query: 487 CSHSGQIERGLRFFR-MIRKYGLDPKPEHFGCVVDLLGRAGQLGEARDLVQELA-EPPAS 544
CSH+G IE G++ F M+ +Y L P EH+G +VDLLGR G+L +A D++ E+ +
Sbjct: 7 CSHAGLIEEGIKMFHVMVNEYQLMPNTEHYGIMVDLLGRMGELDKALDMINEMPMQAGPH 186
Query: 545 VFDSLLGACRCYLDSNLGEEMAMKLIDIEPKNPAPLVVLSNIYAALGRWSEVERIRGLIT 604
V+ +LLGACR + + +GE A+ L ++P + +LSNIY W + ++R LI
Sbjct: 187 VWGALLGACRIHQNIKIGELAALNLFLLDPNHAGYYTLLSNIYCVDKNWHDAAKLRTLIK 366
Query: 605 DKGLDKNSGISMIEV 619
+ K G SM+E+
Sbjct: 367 ENRFKKIVGQSMVEI 411
>BM893152
Length = 356
Score = 98.2 bits (243), Expect = 1e-20
Identities = 48/104 (46%), Positives = 64/104 (61%)
Frame = +3
Query: 411 LATALVDTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNGDYESAFEVFYEMLD 470
L TAL+D Y KCG + +R VFD K D WNAMI GYG NG ESA ++F M +
Sbjct: 51 LGTALIDMYAKCGQLQKSRMVFDSMMEK--DVICWNAMISGYGMNGYAESALDIFQHMEE 224
Query: 471 EMVQPNSATFVSVLSACSHSGQIERGLRFFRMIRKYGLDPKPEH 514
V PN TF+S+LSAC+H+G +E G F ++ Y ++P +H
Sbjct: 225 SNVMPNGITFLSLLSACAHAGLVEEGKYMFARMKSYSVNPNLKH 356
Score = 75.1 bits (183), Expect = 9e-14
Identities = 40/114 (35%), Positives = 68/114 (59%)
Frame = +3
Query: 262 GLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSEKRNLITWNSMIAGMMMNSESE 321
GL MK+ + + T+L+DMY+KCG + VF ++++I WN+MI+G MN +E
Sbjct: 15 GLLMKV-VLPSIPLGTALIDMYAKCGQLQKSRMVFDSMMEKDVICWNAMISGYGMNGYAE 191
Query: 322 RAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFSKMQCAGVAPCLK 375
A+++F+ M + ++P+ T+ SL+S A G+ E F++M+ V P LK
Sbjct: 192 SALDIFQHMEESNVMPNGITFLSLLSACAHAGLVEEGKYMFARMKSYSVNPNLK 353
Score = 62.8 bits (151), Expect = 4e-10
Identities = 31/86 (36%), Positives = 51/86 (59%)
Frame = +3
Query: 173 VSTSLVTAYSKCGVLVSSNKVFENLRVKNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNL 232
+ T+L+ Y+KCG L S VF+++ K+V+ +NA +SG NG+ D+F+ M +
Sbjct: 51 LGTALIDMYAKCGQLQKSRMVFDSMMEKDVICWNAMISGYGMNGYAESALDIFQHMEES- 227
Query: 233 EEKPNKVTLVSVVSACATLSNIRLGK 258
PN +T +S++SACA + GK
Sbjct: 228 NVMPNGITFLSLLSACAHAGLVEEGK 305
Score = 44.7 bits (104), Expect = 1e-04
Identities = 26/73 (35%), Positives = 40/73 (54%)
Frame = +3
Query: 74 TALIASYAANTRSFHYALELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWLFRQIGFWNI 133
TALI YA + +FD M + + +NA++SG NG A+ +F+ + N+
Sbjct: 57 TALIDMYA-KCGQLQKSRMVFDSMMEKDVICWNAMISGYGMNGYAESALDIFQHMEESNV 233
Query: 134 RPNSVTIVSLLSA 146
PN +T +SLLSA
Sbjct: 234 MPNGITFLSLLSA 272
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,458,252
Number of Sequences: 63676
Number of extensions: 432003
Number of successful extensions: 2791
Number of sequences better than 10.0: 173
Number of HSP's better than 10.0 without gapping: 2405
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2663
length of query: 620
length of database: 12,639,632
effective HSP length: 103
effective length of query: 517
effective length of database: 6,081,004
effective search space: 3143879068
effective search space used: 3143879068
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC136286.11


