

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136286.10 + phase: 0
(449 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
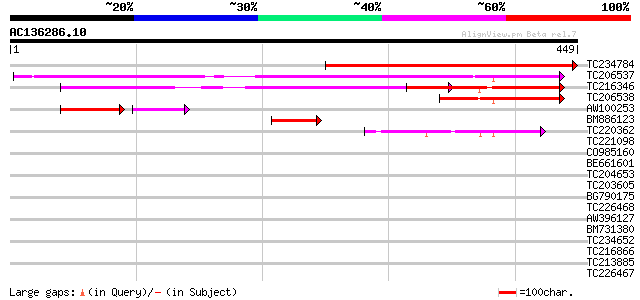
Score E
Sequences producing significant alignments: (bits) Value
TC234784 340 6e-94
TC206537 similar to UP|Q7XYY0 (Q7XYY0) AKIN gamma, complete 278 5e-75
TC216346 similar to UP|Q7XYY0 (Q7XYY0) AKIN gamma, partial (95%) 217 7e-57
TC206538 similar to UP|Q7XYY0 (Q7XYY0) AKIN gamma, partial (27%) 79 4e-15
AW100253 52 3e-11
BM886123 46 3e-05
TC220362 similar to UP|Q41108 (Q41108) Pv42p, partial (58%) 44 1e-04
TC221098 weakly similar to UP|UMP3_ARATH (Q9LEV3) Protein At5g10... 37 0.018
CO985160 34 0.12
BE661601 weakly similar to GP|12483839|gb| p6.9 {Heliocoverpa ar... 34 0.15
TC204653 similar to UP|Q9AT20 (Q9AT20) Histone H1 (Fragment), pa... 33 0.26
TC203605 similar to UP|O49622 (O49622) NAP16kDa protein, partial... 33 0.26
BG790175 similar to PIR|T09854|T098 proline-rich cell wall prote... 32 0.45
TC226468 weakly similar to UP|O65450 (O65450) Glycine-rich prote... 32 0.58
AW396127 32 0.76
BM731380 32 0.76
TC234652 homologue to UP|Q41108 (Q41108) Pv42p, partial (12%) 32 0.76
TC216866 similar to PIR|T00553|T00553 disease resistance respons... 31 0.99
TC213885 weakly similar to UP|Q9VUB7 (Q9VUB7) CG32133-PA, partia... 31 0.99
TC226467 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 31 0.99
>TC234784
Length = 679
Score = 340 bits (873), Expect = 6e-94
Identities = 163/199 (81%), Positives = 186/199 (92%)
Frame = +1
Query: 251 EPGKADIVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNELIL 310
EPG+ DIVNFITQSAV+QGLEGC+GRDWFDCIA + ++DLGLPFMS D+VISIQSNELIL
Sbjct: 13 EPGRPDIVNFITQSAVVQGLEGCKGRDWFDCIAEKCISDLGLPFMSTDEVISIQSNELIL 192
Query: 311 EAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVS 370
EAFK M+DN+IGGLPV+EGP K IVGNLSIRDIR+LLL+PE+F+NFR LTVMDFMKKIVS
Sbjct: 193 EAFKQMKDNRIGGLPVIEGPKKRIVGNLSIRDIRHLLLRPELFTNFRKLTVMDFMKKIVS 372
Query: 371 ASYESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCFITE 430
+S ++GKVT+PITCKPD+TLQ VIHTLASQSIHRIY V+G D+VVGVITLRDVISCF+TE
Sbjct: 373 SSLQTGKVTQPITCKPDSTLQGVIHTLASQSIHRIYVVDGHDEVVGVITLRDVISCFVTE 552
Query: 431 PDYHFDDYYGFAVKEMLNQ 449
P Y+FDDYYGFAVKEMLNQ
Sbjct: 553 PPYNFDDYYGFAVKEMLNQ 609
>TC206537 similar to UP|Q7XYY0 (Q7XYY0) AKIN gamma, complete
Length = 1566
Score = 278 bits (710), Expect = 5e-75
Identities = 152/441 (34%), Positives = 254/441 (57%), Gaps = 5/441 (1%)
Frame = +1
Query: 4 VQNMAHEQEVRTSTQLSKCDRYFETIQSRKKLPQTLQETLTDSFAKIPVSSFPGVPGGKV 63
V MA +E+ S + +K E + ++ + E L F IPVS+FP P
Sbjct: 34 VMAMATMEEIPQSPE-AKLGMRVEDLWDVQEAQLSPTEKLNACFESIPVSAFPLDPSNTE 210
Query: 64 VEILADTPVGEAVKILSESNILAAPVKDPDAGIGSDWRDRYLGIIDYSAIILWVMESAEL 123
+EI +D + +AVKIL+ N+ +APV D +A + W DRY+GI++++ I++W++ +E
Sbjct: 211 IEIKSDATLADAVKILAGHNVFSAPVVDVEAPEDASWIDRYIGIVEFAGIVVWILHQSEP 390
Query: 124 AAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAADKTMAK 183
+ + ++A + A GA A+ L A G L +AA A
Sbjct: 391 TSPRTPSTPSSARAIAAAANGAAFALELEALG------LGSAATTAG------------- 513
Query: 184 DAPQAANNLGEDFYKVILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYR 243
+F++ + E +K+T VR I ++RWAPF+ + ++++ LT+LLLLSKY+
Sbjct: 514 -----------NFFEDLTSSELYKNTKVRDISGTFRWAPFLALERSNSFLTMLLLLSKYK 660
Query: 244 LRNVPVIEPGKADIVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISI 303
+++VPV++ G I N ITQSAVI L C G WF+ + ++++GLP ++ +++I +
Sbjct: 661 MKSVPVLDLGSGAIDNIITQSAVIHMLAECAGLQWFESWGTKKLSEVGLPLVTGNQIIKV 840
Query: 304 QSNELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMD 363
+E +L+AFK+MR ++GG+PV+E K VGN+S+RD+++LL PEI+ ++R +TV D
Sbjct: 841 YEDEPVLQAFKVMRKKRVGGVPVIERETKKAVGNISLRDVQFLLTAPEIYHDYRGITVKD 1020
Query: 364 FMKKIVSASYESGKVTRP-----ITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVI 418
F+ + V + E K P +TCK D T++ +I L + IHR+Y V+ + G+I
Sbjct: 1021FLTE-VRSYLEKNKNASPMLNEYVTCKKDCTIKELIQLLDQEKIHRVYVVDDDGDLQGLI 1197
Query: 419 TLRDVISCFITEPDYHFDDYY 439
TLRD+IS + EP +F D++
Sbjct: 1198TLRDIISRLVHEPRGYFGDFF 1260
>TC216346 similar to UP|Q7XYY0 (Q7XYY0) AKIN gamma, partial (95%)
Length = 1766
Score = 217 bits (553), Expect = 7e-57
Identities = 110/311 (35%), Positives = 185/311 (59%)
Frame = +2
Query: 41 ETLTDSFAKIPVSSFPGVPGGKVVEILADTPVGEAVKILSESNILAAPVKDPDAGIGSDW 100
E F IPVS+FP P + +EI +D + EAVKIL+E NIL+APV D DA + W
Sbjct: 356 EKFNACFESIPVSAFPPPPSNQEIEIKSDATLAEAVKILAEHNILSAPVVDVDAPNDASW 535
Query: 101 RDRYLGIIDYSAIILWVMESAELAAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIA 160
DRY+GI++++ I++W++ +E + +G GA AA+
Sbjct: 536 IDRYIGIVEFAGIVVWILHQSEPTSPRSPSG--------------------GAANTAAVN 655
Query: 161 GLTAAAVGAAVVGGVAADKTMAKDAPQAANNLGEDFYKVILQEEPFKSTTVRSILKSYRW 220
G+T+ A G ++A+ +F++ + + +K+T VR I S+RW
Sbjct: 656 GITSEAFGL-----------------ESASTTSGNFFEDLTSSQLYKNTKVRDISGSFRW 784
Query: 221 APFVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKADIVNFITQSAVIQGLEGCRGRDWFD 280
APF+ + ++++ LT+LLLLSKY+++++PV++ G I N ITQS+VI L C G WF+
Sbjct: 785 APFLALERSNSFLTMLLLLSKYKMKSIPVVDLGAGRIDNIITQSSVIHMLAECAGLQWFE 964
Query: 281 CIAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSI 340
+ ++++GLP ++ + +I + +E +L+AFK+MR +IGGLPV++ + T +GN+S+
Sbjct: 965 SWGTKKLSEVGLPMVTPNHIIKVYEDEPVLQAFKLMRKKRIGGLPVMDRGSSTAIGNISL 1144
Query: 341 RDIRYLLLKPE 351
+D+++LL PE
Sbjct: 1145KDVQFLLTAPE 1177
Score = 95.9 bits (237), Expect = 3e-20
Identities = 48/132 (36%), Positives = 81/132 (61%), Gaps = 7/132 (5%)
Frame = +3
Query: 315 IMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVS---- 370
++ +IGGLPV++ + T +GN+S++D+++LL PEI+ ++R++T DF+ I S
Sbjct: 1167 LLLKKRIGGLPVMDRGSSTAIGNISLKDVQFLLTAPEIYHDYRSITAKDFLTAIRSYLEK 1346
Query: 371 ---ASYESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCF 427
A SG + ITC D T++ +I L + IHR+Y + + G+ITLRD+IS
Sbjct: 1347 HEGAFPMSGDL---ITCNKDCTIKELIQLLDHEKIHRVYVADNDGNLEGLITLRDIISRL 1517
Query: 428 ITEPDYHFDDYY 439
+ EP +F D++
Sbjct: 1518 VHEPRGYFGDFF 1553
>TC206538 similar to UP|Q7XYY0 (Q7XYY0) AKIN gamma, partial (27%)
Length = 644
Score = 79.0 bits (193), Expect = 4e-15
Identities = 39/104 (37%), Positives = 63/104 (60%), Gaps = 5/104 (4%)
Frame = +1
Query: 341 RDIRYLLLKPEIFSNFRNLTVMDFMKKIVSASYESGKVTRP-----ITCKPDATLQSVIH 395
RD+++LL PEI+ ++R +TV +F+ ++ S E K P +TCK D T++ +I
Sbjct: 1 RDVQFLLTAPEIYHDYRGITVKNFLTEVRSY-LEKNKNASPMLSEYVTCKKDCTIKELIQ 177
Query: 396 TLASQSIHRIYTVNGQDQVVGVITLRDVISCFITEPDYHFDDYY 439
L + IHR+Y V + G+ITLRD+IS + EP +F D++
Sbjct: 178 LLDQEKIHRVYVVEDDGDLQGLITLRDIISRLVHEPRGYFGDFF 309
>AW100253
Length = 422
Score = 52.0 bits (123), Expect(2) = 3e-11
Identities = 26/51 (50%), Positives = 33/51 (63%)
Frame = +3
Query: 41 ETLTDSFAKIPVSSFPGVPGGKVVEILADTPVGEAVKILSESNILAAPVKD 91
E F IPVS+FP P + +EI +D + EAVKIL+E NIL+APV D
Sbjct: 102 EKFNACFESIPVSAFPPPPSNQEIEIKSDATLAEAVKILAEHNILSAPVGD 254
Score = 33.9 bits (76), Expect(2) = 3e-11
Identities = 15/46 (32%), Positives = 27/46 (58%), Gaps = 1/46 (2%)
Frame = +1
Query: 98 SDWRDRYLGIIDYSAIILWVMESAE-LAAVALSAGTATAAGVGAGT 142
+ W DRY+GI++++ I +W++ +E + + S G A A V T
Sbjct: 274 ASWIDRYIGIVEFAGIGVWILHQSEPTSPRSPSGGAANTAAVNGIT 411
>BM886123
Length = 165
Score = 46.2 bits (108), Expect = 3e-05
Identities = 19/40 (47%), Positives = 32/40 (79%)
Frame = -3
Query: 208 STTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNV 247
S VR I S+RWAPF+ + ++++ LT+LLLLSKY+++++
Sbjct: 121 SKQVRDISGSFRWAPFLALERSNSFLTMLLLLSKYKMKSI 2
>TC220362 similar to UP|Q41108 (Q41108) Pv42p, partial (58%)
Length = 941
Score = 43.9 bits (102), Expect = 1e-04
Identities = 34/164 (20%), Positives = 75/164 (45%), Gaps = 21/164 (12%)
Frame = +2
Query: 282 IAARPMADLGLPFMSADKVISIQSNELILEAFKIMRDNQIGGLPVVE------------- 328
I +R + DL +++ +I +++A K ++ + +P+V
Sbjct: 347 ILSRSVQDL---LAVTEQIYAITDRTKLVDAIKCLKAAMLNAVPIVRASDVDQDDHKQHI 517
Query: 329 -GPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIVSA-----SYESGKVTRP- 381
G + ++G S D+R + ++ ++ + F + + S+ S ++ R
Sbjct: 518 NGRCRKLIGTFSSTDLRGCHIAT--LKSWLGISALAFTEDVASSPLYTESESDTQINRRE 691
Query: 382 -ITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVI 424
+TC ++ L VI ++ +HR++ V+ Q +VGV++L DVI
Sbjct: 692 LVTCFAESPLSEVIEKAETRHVHRVWMVDHQGLLVGVVSLTDVI 823
>TC221098 weakly similar to UP|UMP3_ARATH (Q9LEV3) Protein At5g10860,
mitochondrial precursor, partial (31%)
Length = 882
Score = 37.0 bits (84), Expect = 0.018
Identities = 34/151 (22%), Positives = 70/151 (45%), Gaps = 8/151 (5%)
Frame = +3
Query: 201 LQEEPFKSTTVRSILK--------SYRWAPFVPVAKNSAMLTVLLLLSKYRLRNVPVIEP 252
+Q++ ++ TV +L S+ W + A++ + ++ + ++ V++P
Sbjct: 114 MQQKGLENVTVSEVLMTKGEENVGSWLWC-----RADDAVVNAMKNMADNNIGSLVVLKP 278
Query: 253 GKADIVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNELILEA 312
I +T+ ++ + GR + M D + +I++ S+ IL+A
Sbjct: 279 EGQHIAGIVTERDCLKKIVA-XGRSPLHTQVGQIMTD-------ENNLITVTSDTNILKA 434
Query: 313 FKIMRDNQIGGLPVVEGPAKTIVGNLSIRDI 343
KIM +N I +PV++G IVG +SI D+
Sbjct: 435 MKIMTENHIRHVPVIDG---KIVGMISIVDV 518
>CO985160
Length = 777
Score = 34.3 bits (77), Expect = 0.12
Identities = 28/73 (38%), Positives = 38/73 (51%), Gaps = 3/73 (4%)
Frame = +2
Query: 113 IILWVMESAELAAVALSAGTATAAGVGAGTVGALG---AIALGATGPAAIAGLTAAAVGA 169
++ V+ + E+AA A +A AA VGA V + A A+ AT A G +A V A
Sbjct: 563 VVAAVVGAEEVAAAAAAAEEVVAAVVGAEEVAVVACEVAAAVVATAEDAAVGFASADVVA 742
Query: 170 AVVGGVAADKTMA 182
A V AAD T+A
Sbjct: 743 AFV--AAADSTIA 775
Score = 31.6 bits (70), Expect = 0.76
Identities = 21/61 (34%), Positives = 29/61 (47%)
Frame = -1
Query: 120 SAELAAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAADK 179
+A ++ A +A T ++A A T + A ATGPAA A A A +A AA
Sbjct: 573 AATTSSAAAAAATTSSAAAAAATTSSAATAA--ATGPAAAACAAATAAASANPATAAATS 400
Query: 180 T 180
T
Sbjct: 399 T 397
Score = 30.4 bits (67), Expect = 1.7
Identities = 21/70 (30%), Positives = 27/70 (38%)
Frame = -1
Query: 120 SAELAAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAADK 179
SA AA A S+ TAA + A + A + TAAA G A AA
Sbjct: 618 SAAAAAAATSSAPTTAATTSSAAAAAATTSSAAAAAATTSSAATAAATGPAAAACAAATA 439
Query: 180 TMAKDAPQAA 189
+ + AA
Sbjct: 438 AASANPATAA 409
>BE661601 weakly similar to GP|12483839|gb| p6.9 {Heliocoverpa armigera
nucleopolyhedrovirus G4}, partial (56%)
Length = 658
Score = 33.9 bits (76), Expect = 0.15
Identities = 30/110 (27%), Positives = 46/110 (41%)
Frame = -3
Query: 84 ILAAPVKDPDAGIGSDWRDRYLGIIDYSAIILWVMESAELAAVALSAGTATAAGVGAGTV 143
++ V +P+ G+G ++G + +S ++ E+ A AG A AA G G V
Sbjct: 359 VVVVVVANPNLGVG------FVGTVCFSGVVA----GEEIGEGAAGAGGAGAAAGGGGGV 210
Query: 144 GALGAIALGATGPAAIAGLTAAAVGAAVVGGVAADKTMAKDAPQAANNLG 193
G GA LGA G VVGG A + + +A + G
Sbjct: 209 GGAGA-RLGAA---------EVGQGRGVVGGDAGEAFTEAEGAEAGDGDG 90
>TC204653 similar to UP|Q9AT20 (Q9AT20) Histone H1 (Fragment), partial (68%)
Length = 1501
Score = 33.1 bits (74), Expect = 0.26
Identities = 36/123 (29%), Positives = 52/123 (42%), Gaps = 2/123 (1%)
Frame = -3
Query: 55 FPGVPGGKVVEILADTPVGEAVKILSESNILAAPVKDPDAGIGSDWRDRYLGIIDYSAII 114
FPG V+E LA +G A + + + +LAA + G+G LG+
Sbjct: 878 FPGEVRVDVLEALAGFALGFAAGLAAAAFVLAAGL-----GLGLAAGLVPLGL------- 735
Query: 115 LWVMESAELAAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGL--TAAAVGAAVV 172
A + L A A A G+GAG +G G +ALG +AG A V A+
Sbjct: 734 -------AAAGLGLEAAAAAAFGLGAG-LGLAGTLALGLAAALGLAGAFDLVALVSLALA 579
Query: 173 GGV 175
G+
Sbjct: 578 AGL 570
>TC203605 similar to UP|O49622 (O49622) NAP16kDa protein, partial (52%)
Length = 906
Score = 33.1 bits (74), Expect = 0.26
Identities = 20/45 (44%), Positives = 22/45 (48%)
Frame = -2
Query: 124 AAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVG 168
A + LSAG AG GAG A GA A A G AGL + G
Sbjct: 323 AGICLSAGAGAGAGAGAGAAAAAGAGAAAAAG----AGLCLSTGG 201
>BG790175 similar to PIR|T09854|T098 proline-rich cell wall protein - upland
cotton, partial (21%)
Length = 415
Score = 32.3 bits (72), Expect = 0.45
Identities = 24/70 (34%), Positives = 28/70 (39%), Gaps = 9/70 (12%)
Frame = -3
Query: 117 VMESAELAAVALSAGTATA---------AGVGAGTVGALGAIALGATGPAAIAGLTAAAV 167
+ ESAE A AL A A AGVGA +G +A G +A
Sbjct: 254 IPESAEPAMKALLAENAAEGGGGVAGEEAGVGAAVGVGVGVVAGGDVAGGGVAAAGGGVA 75
Query: 168 GAAVVGGVAA 177
G V GGV A
Sbjct: 74 GGGVAGGVGA 45
>TC226468 weakly similar to UP|O65450 (O65450) Glycine-rich protein, partial
(20%)
Length = 1041
Score = 32.0 bits (71), Expect = 0.58
Identities = 24/54 (44%), Positives = 25/54 (45%), Gaps = 5/54 (9%)
Frame = +1
Query: 130 AGTA-TAAGVGAGT----VGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAAD 178
AGT+ T AG GAGT GA A G TG AG A GA V A D
Sbjct: 445 AGTSITGAGAGAGTSITGAGAGVAAGGGGTGDGVGAGFVGAGAGAGVAFAGAGD 606
>AW396127
Length = 411
Score = 31.6 bits (70), Expect = 0.76
Identities = 14/47 (29%), Positives = 25/47 (52%)
Frame = +3
Query: 381 PITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCF 427
P+ K TL+ ++ + + +N +QV G++TLRD+I F
Sbjct: 93 PVANKETETLKQIMEHTTETNSSFSFLINDNEQVTGLLTLRDIILQF 233
>BM731380
Length = 433
Score = 31.6 bits (70), Expect = 0.76
Identities = 19/55 (34%), Positives = 30/55 (54%), Gaps = 2/55 (3%)
Frame = +1
Query: 124 AAVALSAGTATAAGVGAGTVGALGA--IALGATGPAAIAGLTAAAVGAAVVGGVA 176
+A A+ AG + G G+ + G+ A +A TG A+ G++ GAA+ GVA
Sbjct: 247 SASAVGAGFGSIFGSGSSSFGSASAPGVAPTTTGVASAPGVSPTTAGAALAPGVA 411
>TC234652 homologue to UP|Q41108 (Q41108) Pv42p, partial (12%)
Length = 730
Score = 31.6 bits (70), Expect = 0.76
Identities = 13/35 (37%), Positives = 23/35 (65%)
Frame = +3
Query: 390 LQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVI 424
L VI ++ +HR++ V+ + +VGV++L DVI
Sbjct: 18 LSEVIEKAVTRHVHRVWVVDHEGLLVGVVSLTDVI 122
>TC216866 similar to PIR|T00553|T00553 disease resistance response protein
homolog F12L6.9 - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (36%)
Length = 1401
Score = 31.2 bits (69), Expect = 0.99
Identities = 20/67 (29%), Positives = 28/67 (40%), Gaps = 5/67 (7%)
Frame = -1
Query: 123 LAAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAA-----VVGGVAA 177
+A V+ SAG TAAG G ++ + + +AG AAV VV G
Sbjct: 492 IAGVSSSAGAVTAAGASVSGTGGSTWLSSSSPESSVVAGFMVAAVAGVWPLGKVVAGPTC 313
Query: 178 DKTMAKD 184
D + D
Sbjct: 312 DMVVGAD 292
>TC213885 weakly similar to UP|Q9VUB7 (Q9VUB7) CG32133-PA, partial (4%)
Length = 540
Score = 31.2 bits (69), Expect = 0.99
Identities = 29/77 (37%), Positives = 36/77 (46%), Gaps = 6/77 (7%)
Frame = -2
Query: 112 AIILWVMESAELAAVALSAGT----ATAAGVGAGTVGALGAIA--LGATGPAAIAGLTAA 165
A + V +AE AAVA+ G A AA G A GA+A + AA+A AA
Sbjct: 305 AAVAAVAATAEAAAVAVVVGVVEVEAAAAVAGVAEAAAAGAVAAVVAEEVAAAVAAEEAA 126
Query: 166 AVGAAVVGGVAADKTMA 182
A AA VAA + A
Sbjct: 125 AAAAAEEVVVAACEVAA 75
Score = 29.3 bits (64), Expect = 3.8
Identities = 26/72 (36%), Positives = 34/72 (47%)
Frame = +1
Query: 118 MESAELAAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAA 177
+ESA A A + TAT++ V A T A A + AA A ++AA AA +
Sbjct: 1 VESAAATAPADAKPTATSSAV-ATTAAATSHAATTTSSAAAAAAASSAATAAA-----TS 162
Query: 178 DKTMAKDAPQAA 189
T A AP AA
Sbjct: 163 SATTAATAPAAA 198
>TC226467 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (6%)
Length = 1017
Score = 31.2 bits (69), Expect = 0.99
Identities = 24/44 (54%), Positives = 24/44 (54%)
Frame = -3
Query: 131 GTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGG 174
G T AGVGAG VGA GA A G A AG A VGA GG
Sbjct: 508 GGGTGAGVGAGFVGA-GAGA-GVAFAGAGAGGVLAGVGAIFGGG 383
Score = 28.9 bits (63), Expect = 4.9
Identities = 20/66 (30%), Positives = 26/66 (39%)
Frame = -3
Query: 131 GTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAADKTMAKDAPQAAN 190
G A GAG V G A A G + G+ A VGG + +D AA
Sbjct: 268 GLEAGATFGAGGVTGAGLGATIAGGWDVVGGVLVVGGDATTVGGSVTASGVPEDGDVAAG 89
Query: 191 NLGEDF 196
+G +F
Sbjct: 88 VVGGEF 71
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,207,017
Number of Sequences: 63676
Number of extensions: 170641
Number of successful extensions: 1059
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 1013
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1043
length of query: 449
length of database: 12,639,632
effective HSP length: 100
effective length of query: 349
effective length of database: 6,272,032
effective search space: 2188939168
effective search space used: 2188939168
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC136286.10


