

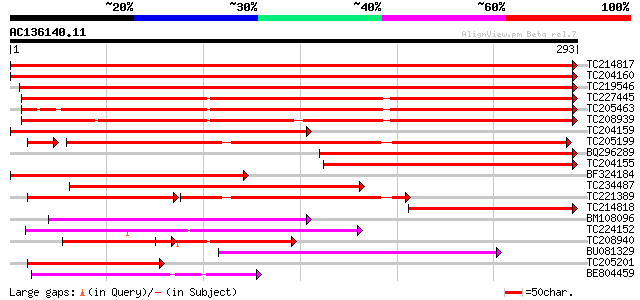
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136140.11 - phase: 0 /pseudo
(293 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, ... 555 e-159
TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate ... 550 e-157
TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete 510 e-145
TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum H... 377 e-105
TC205463 UP|Q39804 (Q39804) BiP isoform B, complete 364 e-101
TC208939 UP|Q39830 (Q39830) BiP isoform A, complete 353 4e-98
TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial ... 277 5e-75
TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa pro... 271 7e-75
BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cyto... 256 6e-69
TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, ... 249 8e-67
BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chapero... 236 9e-63
TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%) 230 5e-61
TC221389 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa p... 122 9e-47
TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous hea... 175 2e-44
BM108096 96 2e-20
TC224152 similar to UP|Q8L8N8 (Q8L8N8) 70kD heat shock protein, ... 95 4e-20
TC208940 UP|Q39803 (Q39803) BiP isoform C (Fragment), partial (26%) 92 3e-19
BU081329 88 5e-18
TC205201 similar to UP|HS7M_PEA (P37900) Heat shock 70 kDa prote... 79 2e-15
BE804459 similar to PIR|T10248|T10 heat shock protein 70K chlo... 70 9e-13
>TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, complete
Length = 2364
Score = 555 bits (1430), Expect = e-159
Identities = 280/293 (95%), Positives = 287/293 (97%)
Frame = +2
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYV FTD+ERLIGDAAKN
Sbjct: 86 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAKN 265
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNP+NTVFDAKRLIGRR SD+SVQSD+KLWPFKVI G +KPMI VNYKGE+KQFAA
Sbjct: 266 QVAMNPINTVFDAKRLIGRRFSDSSVQSDIKLWPFKVIPGAADKPMIVVNYKGEEKQFAA 445
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNV+RIINEP
Sbjct: 446 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEP 625
Query: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD
Sbjct: 626 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 805
Query: 241 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL
Sbjct: 806 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 964
>TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate 70 kDa
protein 2, partial (66%)
Length = 1390
Score = 550 bits (1418), Expect = e-157
Identities = 277/293 (94%), Positives = 285/293 (96%)
Frame = +2
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTD+ERLIGDAAKN
Sbjct: 95 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKN 274
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNP NTVFDAKRLIGRR SDASVQ DMKLWPFKVI GP EKPMI VNYKGE+KQF+A
Sbjct: 275 QVAMNPTNTVFDAKRLIGRRFSDASVQGDMKLWPFKVIPGPAEKPMIVVNYKGEEKQFSA 454
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVL+KM+EIAEAYLGST+KNAVVTVPAYFNDSQRQATKDAGVI+GLNV+RIINEP
Sbjct: 455 EEISSMVLMKMKEIAEAYLGSTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIINEP 634
Query: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
TAAAIAYGLDKKATS GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD
Sbjct: 635 TAAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 814
Query: 241 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
NRMVNHFVQEFKRKNKKDISGN RALRRLRTACERAKRTLSSTAQTTIEIDSL
Sbjct: 815 NRMVNHFVQEFKRKNKKDISGNARALRRLRTACERAKRTLSSTAQTTIEIDSL 973
>TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete
Length = 2099
Score = 510 bits (1314), Expect = e-145
Identities = 257/288 (89%), Positives = 272/288 (94%)
Frame = +1
Query: 6 EGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMN 65
EG AIGIDLGTTYSCVGVWQ+DRVEII NDQGNRTTPSYVAFTD+ERLIGDAAKNQVAMN
Sbjct: 13 EGKAIGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKNQVAMN 192
Query: 66 PVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISS 125
P NTVFDAKRLIGRR SD+SVQ+DMKLWPFKV P +KPMI VNYKGE+K+F+AEEISS
Sbjct: 193 PQNTVFDAKRLIGRRFSDSSVQNDMKLWPFKVGGSPCDKPMIVVNYKGEEKKFSAEEISS 372
Query: 126 MVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 185
MVL+KMRE+AEA+LG VKNAVVTVPAYFNDSQRQATKDAG I+GLNVLRIINEPTAAAI
Sbjct: 373 MVLVKMREVAEAFLGHAVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAAAI 552
Query: 186 AYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVN 245
AYGLDKKA+ GE+NVLIFDLGGGTFDVS+LTIEEGIFEVKATAGDTHLGGEDFDNRMVN
Sbjct: 553 AYGLDKKASRKGEQNVLIFDLGGGTFDVSILTIEEGIFEVKATAGDTHLGGEDFDNRMVN 732
Query: 246 HFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
HFV EFKRKNKKDISGN RALRRLRTACERAKRTLSSTAQTTIEIDSL
Sbjct: 733 HFVSEFKRKNKKDISGNARALRRLRTACERAKRTLSSTAQTTIEIDSL 876
>TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum HSC70-cognate
binding protein precursor, complete
Length = 2254
Score = 377 bits (968), Expect = e-105
Identities = 196/288 (68%), Positives = 231/288 (80%), Gaps = 1/288 (0%)
Frame = +2
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+++ VEIIANDQGNR TPS+VAFTDSERLIG+AAKN A+NP
Sbjct: 209 GTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVAFTDSERLIGEAAKNLAAVNP 388
Query: 67 VNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYK-GEDKQFAAEEISS 125
T+FD KRLIGR+ D VQ DMKL P+K++ G KP I V K GE K F+ EEIS+
Sbjct: 389 ERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNKDG-KPYIQVKIKDGETKVFSPEEISA 565
Query: 126 MVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 185
M+L KM+E AEA+LG + +AVVTVPAYFND+QRQATKDAGVIAGLNV RIINEPTAAAI
Sbjct: 566 MILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAAI 745
Query: 186 AYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVN 245
AYGLDKK GEKN+L+FDLGGGTFDVS+LTI+ G+FEV AT GDTHLGGEDFD R++
Sbjct: 746 AYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRIME 916
Query: 246 HFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
+F++ K+K+ KDIS + RAL +LR ERAKR LSS Q +EI+SL
Sbjct: 917 YFIKLIKKKHGKDISKDNRALGKLRREAERAKRALSSQHQVRVEIESL 1060
>TC205463 UP|Q39804 (Q39804) BiP isoform B, complete
Length = 2383
Score = 364 bits (934), Expect = e-101
Identities = 196/288 (68%), Positives = 228/288 (79%), Gaps = 1/288 (0%)
Frame = +3
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDL TTYSCVGV D VEIIANDQGNR TPS+VAFTDSERLIG+AAK A+NP
Sbjct: 192 GTVIGIDL-TTYSCVGVT--DAVEIIANDQGNRITPSWVAFTDSERLIGEAAKIVAAVNP 362
Query: 67 VNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYK-GEDKQFAAEEISS 125
V T+FD KRLIGR+ D VQ DMKL P+K++ G KP I V K GE K F+ EEIS+
Sbjct: 363 VRTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNKDG-KPYIQVKIKDGETKVFSPEEISA 539
Query: 126 MVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 185
M+L KM+E AEA+LG + +AVVTVPAYFND+QRQATKDAGVIAGLNV RIINEPTAAAI
Sbjct: 540 MILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAAI 719
Query: 186 AYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVN 245
AYGLDKK GEKN+L+FDLGGGTFDVS+LTI+ G+FEV AT GDTHLGGEDFD R++
Sbjct: 720 AYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRIME 890
Query: 246 HFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
+F++ K+K+ KDIS + RAL +LR ERAKR LSS Q +EI+SL
Sbjct: 891 YFIKLIKKKHGKDISKDNRALGKLRREAERAKRALSSQHQVRVEIESL 1034
>TC208939 UP|Q39830 (Q39830) BiP isoform A, complete
Length = 2452
Score = 353 bits (907), Expect = 4e-98
Identities = 188/288 (65%), Positives = 224/288 (77%), Gaps = 1/288 (0%)
Frame = +3
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+++ VEIIANDQGNR TPS+ +FTDSERLIG+AAKN A+NP
Sbjct: 171 GTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSW-SFTDSERLIGEAAKNLAAVNP 347
Query: 67 VNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYK-GEDKQFAAEEISS 125
+FD KRLIGR+ D VQ DMKL P+K++ G KP I K GE K F+ EEIS+
Sbjct: 348 ERVIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNKDG-KPYIQEKIKDGETKVFSPEEISA 524
Query: 126 MVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 185
M+L KM+E AEA+LG + +AV AYFND+QRQATKDAGVIAGLNV RIINEPTAAAI
Sbjct: 525 MILTKMKETAEAFLGKKINDAV----AYFNDAQRQATKDAGVIAGLNVARIINEPTAAAI 692
Query: 186 AYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVN 245
AYGLDKK GEKN+L+FDLGGGTFDVS+LTI+ G+FEV AT GDTHLGGEDFD R++
Sbjct: 693 AYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRIME 863
Query: 246 HFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
+F++ +K+KKDIS + RAL +LR ERAKR LSS Q +EI+SL
Sbjct: 864 YFIKLINKKHKKDISKDSRALSKLRREAERAKRALSSQHQVRVEIESL 1007
>TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial (22%)
Length = 769
Score = 277 bits (708), Expect = 5e-75
Identities = 138/157 (87%), Positives = 146/157 (92%), Gaps = 1/157 (0%)
Frame = +2
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTD+ERLIGDAAKN
Sbjct: 80 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKN 259
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNP+NTVFDAKRLIGRR SDASVQSDMKLWPFKVI GP +KPMI VNYKG++KQF+A
Sbjct: 260 QVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKVIPGPADKPMIVVNYKGDEKQFSA 439
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVP-AYFND 156
EEISSMVLIKM+EIAEAYLGST+KNA YFND
Sbjct: 440 EEISSMVLIKMKEIAEAYLGSTIKNASCHPSLLYFND 550
>TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (94%)
Length = 2204
Score = 271 bits (692), Expect(2) = 7e-75
Identities = 141/262 (53%), Positives = 183/262 (69%), Gaps = 1/262 (0%)
Frame = +3
Query: 30 EIIANDQGNRTTPSYVAFTD-SERLIGDAAKNQVAMNPVNTVFDAKRLIGRRISDASVQS 88
++I N +G RTTPS VAF +E L+G AK Q NP NT+F KRLIGRR D+ Q
Sbjct: 186 KVIENSEGARTTPSVVAFNQKAELLVGTPAKRQAVTNPTNTLFGTKRLIGRRFDDSQTQK 365
Query: 89 DMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVLIKMREIAEAYLGSTVKNAVV 148
+MK+ P+K++ P + N +Q++ ++ + VL KM+E AE+YLG +V AV+
Sbjct: 366 EMKMVPYKIVKAPNGDAWVEAN----GQQYSPSQVGAFVLTKMKETAESYLGKSVSKAVI 533
Query: 149 TVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYGLDKKATSVGEKNVLIFDLGG 208
TVPAYFND+QRQATKDAG IAGL+V RIINEPTAAA++YG++ K E + +FDLGG
Sbjct: 534 TVPAYFNDAQRQATKDAGRIAGLDVQRIINEPTAAALSYGMNNK-----EGLIAVFDLGG 698
Query: 209 GTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRR 268
GTFDVS+L I G+FEVKAT GDT LGGEDFDN +++ V EFKR D+S + AL+R
Sbjct: 699 GTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRTENIDLSKDKLALQR 878
Query: 269 LRTACERAKRTLSSTAQTTIEI 290
LR A E+AK LSST+QT I +
Sbjct: 879 LREAAEKAKIELSSTSQTEINL 944
Score = 27.7 bits (60), Expect(2) = 7e-75
Identities = 12/16 (75%), Positives = 13/16 (81%)
Frame = +1
Query: 10 IGIDLGTTYSCVGVWQ 25
IGIDLGTT SCV V +
Sbjct: 49 IGIDLGTTNSCVSVME 96
>BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cytosolic
[imported] - spinach, partial (21%)
Length = 421
Score = 256 bits (655), Expect = 6e-69
Identities = 130/133 (97%), Positives = 133/133 (99%)
Frame = +1
Query: 161 ATKDAGVIAGLNVLRIINEPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEE 220
ATKDAGVIAGLNV+RIINEPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEE
Sbjct: 1 ATKDAGVIAGLNVMRIINEPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEE 180
Query: 221 GIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTL 280
GIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRK+KKDI+GNPRALRRLRTACERAKRTL
Sbjct: 181 GIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKHKKDINGNPRALRRLRTACERAKRTL 360
Query: 281 SSTAQTTIEIDSL 293
SSTAQTTIEIDSL
Sbjct: 361 SSTAQTTIEIDSL 399
>TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, partial
(75%)
Length = 1640
Score = 249 bits (637), Expect = 8e-67
Identities = 127/131 (96%), Positives = 129/131 (97%)
Frame = +2
Query: 163 KDAGVIAGLNVLRIINEPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGI 222
KDAGVI+GLNV+RIINEPTAAAIAYGLDKKATS GEKNVLIFDLGGGTFDVSLLTIEEGI
Sbjct: 2 KDAGVISGLNVMRIINEPTAAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGI 181
Query: 223 FEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSS 282
FEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGN RALRRLRTACERAKRTLSS
Sbjct: 182 FEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNARALRRLRTACERAKRTLSS 361
Query: 283 TAQTTIEIDSL 293
TAQTTIEIDSL
Sbjct: 362 TAQTTIEIDSL 394
>BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chaperone hsp70 -
rice (fragment), partial (18%)
Length = 398
Score = 236 bits (602), Expect = 9e-63
Identities = 113/123 (91%), Positives = 119/123 (95%)
Frame = -3
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYV FTD+ERLIGDAAKN
Sbjct: 369 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAKN 190
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNP+NTVFDAKRLIGRR SDASVQSD+KLWPFKV++GP EKPMI V+YKGEDKQFAA
Sbjct: 189 QVAMNPINTVFDAKRLIGRRFSDASVQSDIKLWPFKVLSGPAEKPMIQVSYKGEDKQFAA 10
Query: 121 EEI 123
EEI
Sbjct: 9 EEI 1
>TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%)
Length = 457
Score = 230 bits (587), Expect = 5e-61
Identities = 115/152 (75%), Positives = 129/152 (84%)
Frame = +1
Query: 32 IANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVNTVFDAKRLIGRRISDASVQSDMK 91
I NDQGN TTPS VAFTD +RLIG+AAKNQ A NP NTVFDAKRLIGR+ SD +Q D
Sbjct: 1 IHNDQGNNTTPSCVAFTDQQRLIGEAAKNQAATNPENTVFDAKRLIGRKFSDPVIQKDKM 180
Query: 92 LWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVLIKMREIAEAYLGSTVKNAVVTVP 151
LWPFKV+AG +KPMI +NYKG++K AEE+SSMVLIKMREIAEAYL + V+NAVVTVP
Sbjct: 181 LWPFKVVAGINDKPMISLNYKGQEKHLLAEEVSSMVLIKMREIAEAYLETPVENAVVTVP 360
Query: 152 AYFNDSQRQATKDAGVIAGLNVLRIINEPTAA 183
AYFNDSQR+AT DAG IAGLNV+RIINEPTAA
Sbjct: 361 AYFNDSQRKATIDAGAIAGLNVMRIINEPTAA 456
>TC221389 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa protein,
mitochondrial precursor, partial (36%)
Length = 827
Score = 122 bits (305), Expect(2) = 9e-47
Identities = 63/119 (52%), Positives = 84/119 (69%)
Frame = +1
Query: 89 DMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVLIKMREIAEAYLGSTVKNAVV 148
+MK+ PFK++ P + N +Q++ +I + VL KM+E AEAYLG ++ AV+
Sbjct: 496 EMKMVPFKIVKAPNGNAWVETN----GQQYSPSQIGAFVLTKMKETAEAYLGKSISKAVI 663
Query: 149 TVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYGLDKKATSVGEKNVLIFDLG 207
TVPAYFND+QRQATKDAG IAGL+V RIINEPTAAA++YG++KK E + FDLG
Sbjct: 664 TVPAYFNDAQRQATKDAGRIAGLDVQRIINEPTAAALSYGMNKK-----EGLIAXFDLG 825
Score = 82.8 bits (203), Expect(2) = 9e-47
Identities = 43/79 (54%), Positives = 49/79 (61%), Gaps = 1/79 (1%)
Frame = +3
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTD-SERLIGDAAKNQVAMNPVN 68
IGIDLGTT SCV V + ++I N +G RTTPS VAF E L+G K Q NP N
Sbjct: 255 IGIDLGTTNSCVSVMEGKNPKVIENSEGARTTPSVVAFNQKGELLVGTPXKRQAVTNPTN 434
Query: 69 TVFDAKRLIGRRISDASVQ 87
T+F KRLIGRR DA Q
Sbjct: 435 TLFGTKRLIGRRFDDAQTQ 491
>TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat shock
cognate protein 70, partial (68%)
Length = 1554
Score = 175 bits (443), Expect = 2e-44
Identities = 87/87 (100%), Positives = 87/87 (100%)
Frame = +2
Query: 207 GGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRAL 266
GGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRAL
Sbjct: 8 GGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRAL 187
Query: 267 RRLRTACERAKRTLSSTAQTTIEIDSL 293
RRLRTACERAKRTLSSTAQTTIEIDSL
Sbjct: 188 RRLRTACERAKRTLSSTAQTTIEIDSL 268
>BM108096
Length = 525
Score = 95.9 bits (237), Expect = 2e-20
Identities = 50/137 (36%), Positives = 73/137 (52%), Gaps = 1/137 (0%)
Frame = +3
Query: 21 VGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVNTVFDAKRLIGRR 80
V V + ++++ ND+ R TP+ V F D +R +G A MNP N++ KRLIGR+
Sbjct: 9 VAVARQRGIDVVLNDESKRETPAIVCFGDKQRFLGTAGAASTMMNPKNSISQIKRLIGRQ 188
Query: 81 ISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVLIKMREIAEAYLG 140
SD +Q D+K +PF V GP P+I Y GE + F ++ M+L ++EIA L
Sbjct: 189 FSDPELQRDLKTFPFVVTEGPDGYPLIXARYLGEARTFTPTQVFGMMLSNLKEIAXKNLE 368
Query: 141 -STVKNAVVTVPAYFND 156
A +P YF D
Sbjct: 369 CXRCLIACXGIPLYFTD 419
>TC224152 similar to UP|Q8L8N8 (Q8L8N8) 70kD heat shock protein, partial
(33%)
Length = 563
Score = 94.7 bits (234), Expect = 4e-20
Identities = 63/177 (35%), Positives = 98/177 (54%), Gaps = 3/177 (1%)
Frame = +1
Query: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 66
AIGID+GT+ V VW +VE++ N + + SYV F D+ G +++ ++ M
Sbjct: 34 AIGIDIGTSQCSVAVWNGSQVELLKNTRNQKIMKSYVTFKDNIPSGGVSSQLSHEDEMLS 213
Query: 67 VNTVFDAKRLIGRRISDASVQSDMKLWPFKV-IAGPGEKPMIGVNYKGEDKQFAAEEISS 125
T+F+ KRLIGR +D V + L PF V G +P I + EE+ +
Sbjct: 214 GATIFNMKRLIGRVDTDPVVHACKNL-PFLVQTLDIGVRPFIAALVNNMWRSTTPEEVLA 390
Query: 126 MVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTA 182
+ L+++R +AEA L ++N V+TVP F+ Q + A +AGL+VLR++ EPTA
Sbjct: 391 IFLVELRAMAEAQLKRRIRNVVLTVPVSFSRFQLTRIERACAMAGLHVLRLMPEPTA 561
>TC208940 UP|Q39803 (Q39803) BiP isoform C (Fragment), partial (26%)
Length = 447
Score = 92.0 bits (227), Expect = 3e-19
Identities = 43/59 (72%), Positives = 50/59 (83%)
Frame = +1
Query: 28 RVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVNTVFDAKRLIGRRISDASV 86
+VEIIANDQGNR TPS+VAFTDSERLIG+AAKNQ A+NP T+FD KRLIGR+ D +
Sbjct: 82 QVEIIANDQGNRITPSWVAFTDSERLIGEAAKNQAAVNPERTIFDVKRLIGRKFEDKEI 258
Score = 64.3 bits (155), Expect = 6e-11
Identities = 40/79 (50%), Positives = 52/79 (65%), Gaps = 6/79 (7%)
Frame = +2
Query: 76 LIGRRISDAS-----VQSDMKLWPFKVIAGPGEKPMIGVNYK-GEDKQFAAEEISSMVLI 129
L+ R +S+ S Q DMKL P+K++ G KP I V K GE K F+ EEIS+MVLI
Sbjct: 212 LMSRDLSEESSKIRKYQKDMKLVPYKIVNKDG-KPYIQVKIKDGETKVFSPEEISAMVLI 388
Query: 130 KMREIAEAYLGSTVKNAVV 148
KM+E AEA+LG + +AVV
Sbjct: 389 KMKETAEAFLGKKINDAVV 445
>BU081329
Length = 454
Score = 87.8 bits (216), Expect = 5e-18
Identities = 50/147 (34%), Positives = 78/147 (53%), Gaps = 1/147 (0%)
Frame = +3
Query: 109 VNYKGEDKQFAAEEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVI 168
+ Y E F +I +M+ ++ IAE G+ V + V+ VP+YF + QRQA DA I
Sbjct: 3 LKYLKEIHAFTPVQIVAMLFAHLKTIAEKDFGTAVSDCVIGVPSYFTNLQRQAYLDAAAI 182
Query: 169 AGLNVLRIINEPTAAAIAYGLDK-KATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKA 227
GLN L +I++ TA ++YG+ K + V D+G VS+ + G ++ +
Sbjct: 183 VGLNSLLLIHDCTATGLSYGVYKTDIPNAAHIYVAFVDIGHCDTQVSIAAFQAGQMKILS 362
Query: 228 TAGDTHLGGEDFDNRMVNHFVQEFKRK 254
A D+ LGG DFD + +HF FK +
Sbjct: 363 HAFDSSLGGRDFDEVLFSHFAARFKEQ 443
>TC205201 similar to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (18%)
Length = 467
Score = 79.0 bits (193), Expect = 2e-15
Identities = 41/72 (56%), Positives = 48/72 (65%), Gaps = 1/72 (1%)
Frame = +1
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTD-SERLIGDAAKNQVAMNPVN 68
IGIDLGTT SCV V + ++I N +G RTTPS VAF +E L+G AK Q NP N
Sbjct: 247 IGIDLGTTNSCVSVMEGKNPKVIENSEGARTTPSVVAFNQKAELLVGTPAKRQAVTNPTN 426
Query: 69 TVFDAKRLIGRR 80
T+F KRLIGRR
Sbjct: 427 TLFGTKRLIGRR 462
>BE804459 similar to PIR|T10248|T10 heat shock protein 70K chloroplast -
cucumber, partial (18%)
Length = 478
Score = 70.5 bits (171), Expect = 9e-13
Identities = 46/120 (38%), Positives = 71/120 (58%), Gaps = 1/120 (0%)
Frame = +2
Query: 12 IDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDS-ERLIGDAAKNQVAMNPVNTV 70
IDLGTT V + + II+N +G +TTP A+T++ +RL+G AK QV +NP NT+
Sbjct: 116 IDLGTTNCSVAAFVGGKPTIISNSRGQKTTPFVRAYTNNGDRLVGLIAKRQVVVNPENTL 295
Query: 71 FDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVLIK 130
F +R IG ++S+ V + K ++VI + + ++ KQ AA+EIS+ VL K
Sbjct: 296 FFVERFIGPKMSE--VDEESKHVSYRVIR--DDNGNVKLDCPAIGKQCAADEISAQVLTK 463
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.134 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,895,106
Number of Sequences: 63676
Number of extensions: 88877
Number of successful extensions: 417
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 394
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 396
length of query: 293
length of database: 12,639,632
effective HSP length: 97
effective length of query: 196
effective length of database: 6,463,060
effective search space: 1266759760
effective search space used: 1266759760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC136140.11


