

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
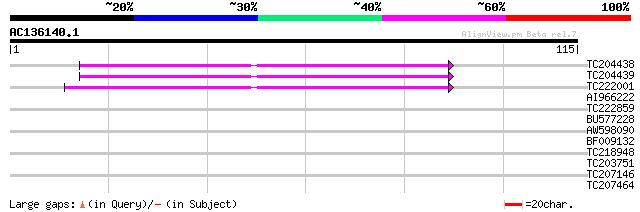
Query= AC136140.1 - phase: 0
(115 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 50 2e-07
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 50 2e-07
TC222001 similar to UP|C716_NEPRA (O04164) Cytochrome P450 71A6 ... 45 4e-06
AI966222 27 1.8
TC222859 homologue to GB|AAO64832.1|29028906|BT005897 At2g02080 ... 25 4.1
BU577228 homologue to GP|4185794|gb|A cysteine desulfurase {Homo... 25 5.4
AW598090 25 5.4
BF009132 similar to GP|3603295|gb| S-adenosyl-methionine cycloar... 25 7.0
TC218948 similar to UP|Q6Q8A0 (Q6Q8A0) Hexokinase 6 , partial (... 24 9.1
TC203751 similar to GB|AAM19926.1|20453375|AY097410 AT3g46430/F1... 24 9.1
TC207146 weakly similar to UP|Q9H4D1 (Q9H4D1) Protein kinase, pa... 24 9.1
TC207464 similar to UP|Q9SGP6 (Q9SGP6) F3M18.8 (At1g28480), part... 24 9.1
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 49.7 bits (117), Expect = 2e-07
Identities = 28/76 (36%), Positives = 41/76 (53%)
Frame = +1
Query: 15 STPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYRFFD 74
+T YE W +KP V F IFG Y QR+K +D S+ + LG S S+AYR F+
Sbjct: 2710 TTLYEIWKGRKPTVKHFHIFGSPCYILADREQRRK-MDPKSDAGIFLGYSTNSRAYRVFN 2886
Query: 75 RIIKKIVVSEDVAFDE 90
+ ++ S +V D+
Sbjct: 2887 SRTRTVMESINVVVDD 2934
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 49.7 bits (117), Expect = 2e-07
Identities = 28/76 (36%), Positives = 41/76 (53%)
Frame = +1
Query: 15 STPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYRFFD 74
+T YE W +KP V F IFG Y QR+K +D S+ + LG S S+AYR F+
Sbjct: 2707 TTLYEIWKGRKPSVKHFHIFGSPCYILADREQRRK-MDPKSDAGIFLGYSTNSRAYRVFN 2883
Query: 75 RIIKKIVVSEDVAFDE 90
+ ++ S +V D+
Sbjct: 2884 SRTRTVMESINVVVDD 2931
>TC222001 similar to UP|C716_NEPRA (O04164) Cytochrome P450 71A6 (Fragment)
, partial (21%)
Length = 912
Score = 45.4 bits (106), Expect = 4e-06
Identities = 24/79 (30%), Positives = 41/79 (51%)
Frame = -2
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
+++++PYE P + RIFG + YA + RKK L+ ++ C+ +G +K Y
Sbjct: 839 LQNTSPYERLHGHIPDISHLRIFGCLCYASTIKANRKK-LEPRAHPCIFIGFKPNTKGYM 663
Query: 72 FFDRIIKKIVVSEDVAFDE 90
+D I+ S +V F E
Sbjct: 662 LYDLHSHNIITSRNVVFYE 606
>AI966222
Length = 430
Score = 26.6 bits (57), Expect = 1.8
Identities = 13/25 (52%), Positives = 14/25 (56%), Gaps = 3/25 (12%)
Frame = +1
Query: 16 TPYEAWSVKKP---YVMPFRIFGFI 37
TPYE W +KP Y PFR FI
Sbjct: 118 TPYELWKGRKPNISYFYPFRCKCFI 192
>TC222859 homologue to GB|AAO64832.1|29028906|BT005897 At2g02080 {Arabidopsis
thaliana;} , partial (22%)
Length = 454
Score = 25.4 bits (54), Expect = 4.1
Identities = 11/24 (45%), Positives = 18/24 (74%), Gaps = 1/24 (4%)
Frame = -2
Query: 38 AYAHI-PFSQRKKTLDDNSNKCVL 60
AYAH+ PF +RKK + +NS + ++
Sbjct: 108 AYAHMFPFQERKKNILNNSKQQII 37
>BU577228 homologue to GP|4185794|gb|A cysteine desulfurase {Homo sapiens},
partial (2%)
Length = 428
Score = 25.0 bits (53), Expect = 5.4
Identities = 22/72 (30%), Positives = 35/72 (48%), Gaps = 1/72 (1%)
Frame = -2
Query: 7 VSSLIVKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEK 66
+S+L+V ST + +++ PY IFG + I F K +D +S L EK
Sbjct: 331 LSTLLVTFSTTFSLFALLDPYSPSRPIFGCVRAVIISF----KIMDVDSGGIKLPCRLEK 164
Query: 67 S-KAYRFFDRII 77
S + RFF ++
Sbjct: 163 SPHSLRFFCNLL 128
>AW598090
Length = 409
Score = 25.0 bits (53), Expect = 5.4
Identities = 16/40 (40%), Positives = 24/40 (60%)
Frame = +3
Query: 68 KAYRFFDRIIKKIVVSEDVAFDEQGMLSLILKEIQIRIYR 107
K R DR + K +S+ F Q M SLILKE+ ++++R
Sbjct: 60 KDKRNIDRQVLKDSLSQLAKF-RQSM*SLILKELIVQVWR 176
>BF009132 similar to GP|3603295|gb| S-adenosyl-methionine
cycloartenol-C24-methyltransferase {Nicotiana tabacum},
partial (37%)
Length = 420
Score = 24.6 bits (52), Expect = 7.0
Identities = 10/23 (43%), Positives = 15/23 (64%)
Frame = -2
Query: 27 YVMPFRIFGFIAYAHIPFSQRKK 49
Y++ F I G +++H PF RKK
Sbjct: 134 YLLVFFIVGIESFSHAPFICRKK 66
>TC218948 similar to UP|Q6Q8A0 (Q6Q8A0) Hexokinase 6 , partial (43%)
Length = 984
Score = 24.3 bits (51), Expect = 9.1
Identities = 9/29 (31%), Positives = 17/29 (58%)
Frame = -2
Query: 5 LWVSSLIVKSSTPYEAWSVKKPYVMPFRI 33
+W+ +L ++ + +W V+ P V PF I
Sbjct: 92 IWIWALSIQVNVIRRSW*VRGPKVSPFHI 6
>TC203751 similar to GB|AAM19926.1|20453375|AY097410 AT3g46430/F18L15_150
{Arabidopsis thaliana;} , partial (80%)
Length = 416
Score = 24.3 bits (51), Expect = 9.1
Identities = 14/42 (33%), Positives = 20/42 (47%), Gaps = 4/42 (9%)
Frame = +1
Query: 20 AWSV----KKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNK 57
+WS+ P P GF+ PFSQR+K NS++
Sbjct: 88 SWSLC*RPSPPPSSPTSKIGFLF*V*FPFSQRRKPQSQNSSR 213
>TC207146 weakly similar to UP|Q9H4D1 (Q9H4D1) Protein kinase, partial (4%)
Length = 1166
Score = 24.3 bits (51), Expect = 9.1
Identities = 15/47 (31%), Positives = 24/47 (50%), Gaps = 2/47 (4%)
Frame = -1
Query: 11 IVKSSTPYEAWSVKKPYVMPFRIFGF--IAYAHIPFSQRKKTLDDNS 55
+VK +T + A + F I GF ++ AHIPFS + D ++
Sbjct: 926 VVKGATIFLAIKETAIQIYCFSI*GFDLVSLAHIPFSSLPSSTDTDT 786
>TC207464 similar to UP|Q9SGP6 (Q9SGP6) F3M18.8 (At1g28480), partial (56%)
Length = 843
Score = 24.3 bits (51), Expect = 9.1
Identities = 11/48 (22%), Positives = 23/48 (47%)
Frame = +1
Query: 9 SLIVKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSN 56
S+I+ + PY +W F IA++H + + +++N+N
Sbjct: 40 SIIMHQAIPYRSWRPLHNPTTHFSPLPLIAHSHNDDDKMRNIVNNNNN 183
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.142 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,820,209
Number of Sequences: 63676
Number of extensions: 52769
Number of successful extensions: 286
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 286
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 286
length of query: 115
length of database: 12,639,632
effective HSP length: 91
effective length of query: 24
effective length of database: 6,845,116
effective search space: 164282784
effective search space used: 164282784
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 51 (24.3 bits)
Medicago: description of AC136140.1


