

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135959.8 + phase: 0 /pseudo/partial
(1454 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
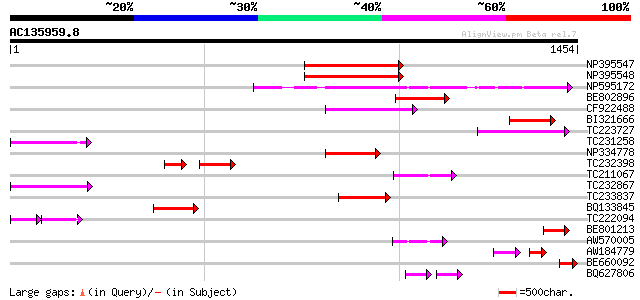
Score E
Sequences producing significant alignments: (bits) Value
NP395547 reverse transcriptase [Glycine max] 401 e-111
NP395548 reverse transcriptase [Glycine max] 386 e-107
NP595172 polyprotein [Glycine max] 366 e-101
BE802896 190 3e-48
CF922488 184 3e-46
BI321666 164 2e-40
TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 136 8e-32
TC231258 similar to UP|Q886T6 (Q886T6) Sensory box histidine kin... 125 1e-28
NP334778 reverse transcriptase [Glycine max] 124 3e-28
TC232398 82 7e-28
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 122 2e-27
TC232867 similar to UP|Q9MA68 (Q9MA68) F2J6.13 protein, partial ... 114 2e-25
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 112 2e-24
BQ133845 101 2e-21
TC222094 60 3e-20
BE801213 weakly similar to GP|6691193|gb| F7F22.17 {Arabidopsis ... 89 2e-17
AW570005 86 2e-16
AW184779 55 1e-14
BE660092 weakly similar to GP|9884624|dbj retroelement pol polyp... 71 3e-12
BQ627806 46 1e-11
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 401 bits (1030), Expect = e-111
Identities = 185/254 (72%), Positives = 218/254 (84%)
Frame = +1
Query: 755 VKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDY 814
V+KEV KLL+AG+IYPISDS WVSPVQVVPKKGG+TV+KND+NE I TR VT WRMCIDY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 815 RKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGT 874
RKLN+ATRKDH+PLPF+DQML+RLA+ S + +LDGYSG+ QI + P DQEKT FTCPF
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 875 FAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLER 934
FAYRRMPFGLCNA TFQRCMM+IF D VEK +EVFMDDFS G++F +CL NLEKVL+R
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 935 CEQVNLVLNWEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLPPTSVKEIRSFLGH 994
CE+ NLVLNWEKCHFMV+EGIVLGH +S RGIEV + K+++I+K+ PP +VK I SFLGH
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 995 AGFYRRFIKDFSSI 1008
GFYRRFIKDF+ +
Sbjct: 721 VGFYRRFIKDFTKV 762
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 386 bits (992), Expect = e-107
Identities = 178/254 (70%), Positives = 214/254 (84%)
Frame = +1
Query: 755 VKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDY 814
V+KEVLKLL+ G+IYPISDS WVSPV VV KK G+TVI+N+KN+ I TRTVT W++CIDY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 815 RKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGT 874
RKLN+ATRKDHFPLPF+DQMLERLA H+++C+LD Y G+ QI + P DQEK FTCPFG
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 875 FAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLER 934
FAYRR+PFGLCNAP TFQ CM++IF+D VEK +EVFMDDFSV + + CL LE VL+R
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 935 CEQVNLVLNWEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLPPTSVKEIRSFLGH 994
C + NLVLNWEKCHFMVREGIVLGH +S RGIEVD+ K+++IEK+ PP++VK IRSFLG
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 995 AGFYRRFIKDFSSI 1008
A FYRRFIKDF+ +
Sbjct: 721 ARFYRRFIKDFTKV 762
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 366 bits (940), Expect = e-101
Identities = 269/829 (32%), Positives = 407/829 (48%), Gaps = 12/829 (1%)
Frame = +1
Query: 626 EGEAKAYEVLLDRTPPIKDL-SVEELIKEEPTLQPKEAPKVELKTLPSNLRYEFLGPNST 684
EG ++A + L +++ S+EE + L KE P+ LK LP+N+ E T
Sbjct: 1531 EGNSEATQAQLHHFRRLQNTKSIEECFAIQ--LIQKEVPEDTLKDLPTNIDPELAILLHT 1704
Query: 685 YPVI--VNASLDEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEDDYKPSIE 742
Y + V ASL + H I L+ P
Sbjct: 1705 YAQVFAVPASLPPQREQD----------------------------HAIPLKQGSGPVKV 1800
Query: 743 HQRRLNPNMKEVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIAT 802
R K+ ++K + ++L G+I P S+S + P+ +V KK G
Sbjct: 1801 RPYRYPHTQKDQIEKMIQEMLVQGIIQP-SNSPFSLPILLVKKKDG-------------- 1935
Query: 803 RTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPND 862
WR C DYR LN T KD FP+P +D++L+ L +F LD SG+ QI + P D
Sbjct: 1936 ----SWRFCTDYRALNAITVKDSFPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQPED 2103
Query: 863 QEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFD 922
+EKT F G + + MPFGL NAPATFQ M IF + K + VF DD ++ +++
Sbjct: 2104 REKTAFRTHHGHYEWLVMPFGLTNAPATFQCLMNKIFQFALRKFVLVFFDDILIYSASWK 2283
Query: 923 DCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLPP 982
D L +LE VL+ +Q L KC F E LGH VS G+ ++ KV+ + P
Sbjct: 2284 DHLKHLESVLQTLKQHQLFARLSKCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDWPTP 2463
Query: 983 TSVKEIRSFLGHAGFYRRFIKDFSSITKPLTNLLLKDADFTFDDSCLQAFCRLKEALITA 1042
+VK++R FLG G+YRRFIK +++I PLT+LL KD+ F +++ AF +LK+A+ A
Sbjct: 2464 NNVKQLRGFLGLTGYYRRFIKSYANIAGPLTDLLQKDS-FLWNNEAEAAFVKLKKAMTEA 2640
Query: 1043 PIIQPPDWNLPFEIMCDASDYAVGVVLGQRKDKKMHAIYYASKTLDGAQVNYATTEKELL 1102
P++ PD++ PF + DAS VG VLGQ H I Y SK L + +ELL
Sbjct: 2641 PVLSLPDFSQPFILETDASGIGVGAVLGQNG----HPIAYFSKKLAPRMQKQSAYTRELL 2808
Query: 1103 AVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGV 1162
A+ A+ KFR YL+G+K I+ TD ++K L+++ P W+ +D +I+ K G
Sbjct: 2809 AITEALSKFRHYLLGNKFIIRTDQRSLKSLMDQSLQTPEQQAWLHKFLGYDFKIEYKPGK 2988
Query: 1163 ENVVADHLSRLREANKDELPLDDSFPDDQLFLLAQTDAPWYADFVNFLAAGVL--PPELN 1220
+N AD LSR +F+LA ++ ++ F+ L A ++ P
Sbjct: 2989 DNQAADALSR-------------------MFMLAWSEP--HSIFLEELRARLISDPHLKQ 3105
Query: 1221 YQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPESE--VSSILTHCHSSSYGGHASTQ 1278
+ K D HY E L+ + R IP E V+ IL HSS GGHA
Sbjct: 3106 LMETYKQGADASHYTVREGLLYWKD-----RVVIPAEEEIVNKILQEYHSSPIGGHAGIT 3270
Query: 1279 KTSFKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITKRNEMPLNNILEVEI-FDVW--- 1334
+T + L + F+WP + +DV +I KC CQ+ S N +P + + I VW
Sbjct: 3271 RTLAR-LKAQFYWPKMQEDVKAYIQKCLICQQAKS---NNTLPAGLLQPLPIPQQVWEDV 3438
Query: 1335 GVDFMGPFPSSFGNQYILVAVDYVSKWVEAIASPTN-DAQVVIKMFKKVIFPRFGVPRVV 1393
+DF+ P+SFG I+V +D ++K+ I + +++VV + F I G+PR +
Sbjct: 3439 AMDFITGLPNSFGLSVIMVVIDRLTKYAHFIPLKADYNSKVVAEAFMSHIVKLHGIPRSI 3618
Query: 1394 ISDGGSHFISKHFEKLLQKLGVRHKVATPYHPQTSGQVEVSNRQIKAIL 1442
+SD F S ++ L + G +++ YHPQ+ GQ EV N+ ++ L
Sbjct: 3619 VSDRDRVFTSTFWQHLFKLQGTTLAMSSAYHPQSDGQSEVLNKCLEMYL 3765
>BE802896
Length = 416
Score = 190 bits (483), Expect = 3e-48
Identities = 89/138 (64%), Positives = 109/138 (78%)
Frame = -2
Query: 990 SFLGHAGFYRRFIKDFSSITKPLTNLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPD 1049
SFLGHAGFYRRFI+DF + PL+NLL K+ +F F+D C AF LK ALIT PIIQ PD
Sbjct: 415 SFLGHAGFYRRFIRDFRKVALPLSNLLQKEVEFDFNDKCK*AFDCLKRALITTPIIQAPD 236
Query: 1050 WNLPFEIMCDASDYAVGVVLGQRKDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAID 1109
W PFE+MCDAS+YA+GVVL Q+ DK IYY+S+TLD AQ NY TTEKELLA+V+A++
Sbjct: 235 WTAPFELMCDASNYALGVVLAQKIDKLPRVIYYSSRTLDAAQANYTTTEKELLAIVFALE 56
Query: 1110 KFRQYLVGSKIIVYTDHS 1127
KF YL+G++IIVY DH+
Sbjct: 55 KFHSYLLGTRIIVYIDHA 2
>CF922488
Length = 741
Score = 184 bits (466), Expect = 3e-46
Identities = 99/236 (41%), Positives = 141/236 (58%)
Frame = +3
Query: 810 MCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFT 869
MC+DYR LN A+ KD FPLP I+ +++ S F ++DG+SG+ QI I P D EKTTF
Sbjct: 30 MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAPEDMEKTTFI 209
Query: 870 CPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLE 929
+GTF Y+ M FGL N AT+QR M+++F D + K +EV+MDD V ++ L NL
Sbjct: 210 TLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRTEEEHLVNLR 389
Query: 930 KVLERCEQVNLVLNWEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLPPTSVKEIR 989
K+ R + L LN KC F V+ +L + S RGIEVD KV++I +M P + K+++
Sbjct: 390 KLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMAKPHTEKQVQ 569
Query: 990 SFLGHAGFYRRFIKDFSSITKPLTNLLLKDADFTFDDSCLQAFCRLKEALITAPII 1045
FLG + RFI + +PL LL K+ +D C AF R+K+ LI ++
Sbjct: 570 GFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLINPHVL 737
>BI321666
Length = 430
Score = 164 bits (416), Expect = 2e-40
Identities = 75/120 (62%), Positives = 94/120 (77%)
Frame = +2
Query: 1281 SFKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITKRNEMPLNNILEVEIFDVWGVDFMG 1340
S +L S F+ PS+FKD ++ C+KCQRT S++KRNE+PL+ ILEVEIFD WG+DF+G
Sbjct: 5 STNVLQSRFYLPSIFKDAYVHAQSCNKCQRTRSVSKRNELPLHTILEVEIFDYWGIDFVG 184
Query: 1341 PFPSSFGNQYILVAVDYVSKWVEAIASPTNDAQVVIKMFKKVIFPRFGVPRVVISDGGSH 1400
PFP SF N+YILV VDYVSKWVEA+A +DA++VIK KK IF R GVP V+I +GGSH
Sbjct: 185 PFPPSFSNEYILVVVDYVSKWVEAVACQKSDAKIVIKFLKKQIFSRLGVPWVLIDNGGSH 364
>TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (9%)
Length = 843
Score = 136 bits (342), Expect = 8e-32
Identities = 71/236 (30%), Positives = 123/236 (52%), Gaps = 2/236 (0%)
Frame = +1
Query: 1201 PWYADFVNFLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPESEVS 1260
PWY D ++ + PE+ K+ ++ L++R D RC+ E +
Sbjct: 136 PWYFDIKRYVISKEYLPEIADNDKRTLRRLAAGFFMSGSILYKRNHDMKPLRCVDAREAN 315
Query: 1261 SILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITKRNEM 1320
++ H S+G HA+ + KIL +G++W ++ D + + KC KCQ
Sbjct: 316 HMIEEVHEGSFGTHANGHAMARKILRAGYYWLTMESDCCVHVRKCHKCQAFADNVNAPPH 495
Query: 1321 PLNNILEVEIFDVWGVDFMGPF--PSSFGNQYILVAVDYVSKWVEAIASPTNDAQVVIKM 1378
PLN + F +WG+D +G +S G+++ILVA+DY +KWVEA + VV++
Sbjct: 496 PLNVMSSPWPFSMWGIDVIGAIEPKASNGHRFILVAIDYFTKWVEAASYTDVMRGVVVRF 675
Query: 1379 FKKVIFPRFGVPRVVISDGGSHFISKHFEKLLQKLGVRHKVATPYHPQTSGQVEVS 1434
KK I R+G+PR +I+D G++ +K ++ ++ ++H TPY P+ + VEV+
Sbjct: 676 IKKEIICRYGLPRKIITDNGTNLNNKMMGEICEEFKIQHHNPTPYRPKMN*AVEVA 843
>TC231258 similar to UP|Q886T6 (Q886T6) Sensory box histidine kinase/response
regulator, partial (3%)
Length = 1374
Score = 125 bits (314), Expect = 1e-28
Identities = 74/210 (35%), Positives = 114/210 (54%), Gaps = 3/210 (1%)
Frame = +2
Query: 2 QFSGSPSEDPNLHISNFWRLSVTCKD---DQETVRLHLFPFSLKNKASNWFNSLKPGSIT 58
+F G EDP+ H+ F + T K ++ + L FP SL+ A +W L P SI
Sbjct: 551 KFHGLVGEDPHKHLKEFHIVCSTMKPPDVQEDHIFLKAFPHSLEGVAKDWLYYLAPRSIF 730
Query: 59 SWDQLRREFLCRFFPPSKTAQLRGKLYQFTQMNGESLFDVWERFKEILRRCPHHGLEKWL 118
+WD L+R FL +FFP S+T +R + Q++ ESL++ WERFK+ CPHH + K L
Sbjct: 731 NWDDLKRVFLEKFFPASRTTTIRKDISGIRQLSRESLYEYWERFKKSCASCPHHQISKQL 910
Query: 119 IIHIFYNGLTSSTKLTIDAAAGGALMNKDFTTAYALIENMALNLFQWTEEKETVDPSPSE 178
++ FY L++ + IDAA+GGAL + T A LI+ MA N Q++ + +
Sbjct: 911 LL*YFYEELSNMKRSMIDAASGGALGDMTPTEARNLIKKMASNSQQFSARNDAI------ 1072
Query: 179 KEAGINETSSIDYLSTKLNALSQRLDRMST 208
G++E ++ ST+ +L + L ST
Sbjct: 1073VLRGVHEVATDSSSSTENKSLRENLMPWST 1162
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 124 bits (311), Expect = 3e-28
Identities = 59/141 (41%), Positives = 89/141 (62%)
Frame = +3
Query: 809 RMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTF 868
RMC+DYR LN+A+ KD+FPLP ID ++ +A + F ++DG+SG+ QI + P D EKTTF
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 869 TCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNL 928
+GTF Y+ M FGL N AT+ R M+++F D + K +E ++D+ ++ L NL
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 929 EKVLERCEQVNLVLNWEKCHF 949
+ + + + L LN KC F
Sbjct: 363 QNLFGQLRKYRLRLNPRKCVF 425
>TC232398
Length = 1054
Score = 82.4 bits (202), Expect(2) = 7e-28
Identities = 38/91 (41%), Positives = 61/91 (66%)
Frame = +1
Query: 487 RLGIGELKSTRMTLQLADRSVKYPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLG 546
R+G +++ TRMTLQLAD S+ G++ED+ VKV ++ DFV+M++EED ++ ++LG
Sbjct: 175 RIGNQKIEPTRMTLQLADHSITRSFGVVEDILVKVHQLIFLVDFVIMDIEEDAEIRLILG 354
Query: 547 RPFLATAGAIIDVKKGKLAFNVGKETVEFEL 577
PF+ TA ++D+ KG L +V + F L
Sbjct: 355 WPFMVTAKCVVDMGKGNLEMSVEDQKATFNL 447
Score = 61.6 bits (148), Expect(2) = 7e-28
Identities = 33/57 (57%), Positives = 39/57 (67%)
Frame = +2
Query: 397 IDVPFTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTEECSAIIQNKLPPKLKDPG 453
I +PF E + QMP Y KFLK+IL KK K SET+ + E C A+IQ KLPPK KD G
Sbjct: 8 ITIPFGERIQQMPLYKKFLKDILIKKGKYINSETIVVGEYCRALIQ-KLPPKFKDLG 175
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 122 bits (305), Expect = 2e-27
Identities = 65/163 (39%), Positives = 96/163 (58%)
Frame = +1
Query: 984 SVKEIRSFLGHAGFYRRFIKDFSSITKPLTNLLLKDADFTFDDSCLQAFCRLKEALITAP 1043
SV +IRSF G A FYRRF+ +FS++ PL L+ K+ FT+ + QAF LKE L AP
Sbjct: 109 SVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKLTKAP 288
Query: 1044 IIQPPDWNLPFEIMCDASDYAVGVVLGQRKDKKMHAIYYASKTLDGAQVNYATTEKELLA 1103
++ PD++ FE+ CDAS V VL Q H I Y S+ L A +NY T +KEL A
Sbjct: 289 VLALPDFSKTFELECDASGVGVRAVLLQGG----HPIAYFSEKLHSATLNYPTYDKELYA 456
Query: 1104 VVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWI 1146
++ A + +LV + ++++DH ++KY+ K R +W+
Sbjct: 457 LIRAPQTWEHFLVCKEFVIHSDHQSLKYIRGKSKLNKRHAKWV 585
>TC232867 similar to UP|Q9MA68 (Q9MA68) F2J6.13 protein, partial (3%)
Length = 1144
Score = 114 bits (286), Expect = 2e-25
Identities = 69/217 (31%), Positives = 113/217 (51%), Gaps = 5/217 (2%)
Frame = -3
Query: 1 NQFSGSPSEDPNLHISNFWRLSVTCK---DDQETVRLHLFPFSLKNKASNWFNSLKPGSI 57
N F G P+EDP H++ + + T + + +RL L FSL +A W +S K S+
Sbjct: 788 NFFHGLPNEDPYAHLATYIEICNTIRLAGVPADAIRLSLLSFSLSGEAKRWLHSFKGNSL 609
Query: 58 TSWDQLRREFLCRFFPPSKTAQLRGKLYQFTQMNGESLFDVWERFKEILRRCPHHGLEKW 117
SWD++ +FL ++FP SKTA+ + + F Q ESL + ERF+ +LR+ P HG +
Sbjct: 608 KSWDEVVEKFLKKYFPESKTAEGKAAISSFHQFPDESLSEALERFRGLLRKTPTHGFSEP 429
Query: 118 LIIHIFYNGLTSSTKLTIDAAAGGALMNKDFTTAYALIENMALNLFQWTEEKETVDPSPS 177
+ ++IF + L +K +DA+ GG + K A LIE+MA + ++ + S
Sbjct: 428 IQLNIFIDELRPESKQLVDASVGGKIKMKTPDEAMDLIESMAASDIAILRDRAHIPTKKS 249
Query: 178 EKEAGINET--SSIDYLSTKLNALSQRLDRMSTPTFS 212
E +T + LS +L L++ L ++ T S
Sbjct: 248 LLELTSQDTLLAQNKLLSKQLETLTKTLSKLPTQLHS 138
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 112 bits (279), Expect = 2e-24
Identities = 58/133 (43%), Positives = 84/133 (62%)
Frame = +2
Query: 844 FCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFV 903
F ++DG+SG+ QI + D EKTTF +GTF+YR M FGL N AT+QR M+++F D +
Sbjct: 2 FSFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMM 181
Query: 904 EKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVSD 963
K +EV++DD + L NL K+ R ++ L LN KC F V+ G +LG +VS
Sbjct: 182 HKEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQ 361
Query: 964 RGIEVDRAKVEII 976
+GIE+D KV+ +
Sbjct: 362 KGIEIDPEKVKAL 400
>BQ133845
Length = 389
Score = 101 bits (252), Expect = 2e-21
Identities = 50/116 (43%), Positives = 74/116 (63%)
Frame = +2
Query: 369 KIPFPQRFAKSKLDEQFKKFMEMMNKIYIDVPFTEVLTQMPTYAKFLKEILSKKRKIEES 428
++P+P + ++ KF+++ K+ I +PF E L QMP YA FLK++L+KK S
Sbjct: 32 EVPYPLVPS*KDKEQHLAKFLDIFKKLEITLPFEEALQQMPLYANFLKDMLTKKNWYIHS 211
Query: 429 ETVNLTEECSAIIQNKLPPKLKDPGSFSIPCVIGSEVVKKAMCDLGASVSLMPLSL 484
+ + + CSA+IQ LPP DPG ++PC IG V KA+ DLGAS++LMPLS+
Sbjct: 212 DKIVVEGNCSAVIQRILPP*HTDPGFVTMPCSIGEVAVGKALIDLGASINLMPLSM 379
>TC222094
Length = 984
Score = 60.1 bits (144), Expect(2) = 3e-20
Identities = 29/85 (34%), Positives = 47/85 (55%), Gaps = 3/85 (3%)
Frame = -2
Query: 1 NQFSGSPSEDPNLHISNFWRLSVTCKD---DQETVRLHLFPFSLKNKASNWFNSLKPGSI 57
N F G P+EDP H++ + + T K ++ + L LF FSL +A W S K ++
Sbjct: 728 NLFYGLPTEDPYAHLATYIDICNTVKIVGVPEDAIHLDLFCFSLAGEARTWLRSFKGNNL 549
Query: 58 TSWDQLRREFLCRFFPPSKTAQLRG 82
+W++ +FL ++FP SKT +G
Sbjct: 548 RTWNEXXEKFLKKYFPESKTIXXKG 474
Score = 58.2 bits (139), Expect(2) = 3e-20
Identities = 35/103 (33%), Positives = 56/103 (53%)
Frame = -3
Query: 83 KLYQFTQMNGESLFDVWERFKEILRRCPHHGLEKWLIIHIFYNGLTSSTKLTIDAAAGGA 142
++ F Q ESL + +RF +L + P HG + + ++IF +G+ +K +DA+AGG
Sbjct: 472 EISSFHQHPHESLSEALDRFHGLLWKTPTHGFSEPVQLNIFIDGMQPHSKQLLDASAGGK 293
Query: 143 LMNKDFTTAYALIENMALNLFQWTEEKETVDPSPSEKEAGINE 185
+ K A LIENMA N + ++E PSP E+ + E
Sbjct: 292 IKLKTPEEAIELIENMAANDYVILRDQE---PSPQEESTRLEE 173
>BE801213 weakly similar to GP|6691193|gb| F7F22.17 {Arabidopsis thaliana},
partial (3%)
Length = 416
Score = 88.6 bits (218), Expect = 2e-17
Identities = 41/66 (62%), Positives = 50/66 (75%)
Frame = +3
Query: 1370 NDAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISKHFEKLLQKLGVRHKVATPYHPQTSG 1429
NDA++VIK KK IF RFG+PR++ISDGGSHF K+L+ VRHKV T YHPQT+G
Sbjct: 3 NDAKIVIKFLKKNIFSRFGMPRILISDGGSHFYYSQLNKVLKHDSVRHKVETSYHPQTNG 182
Query: 1430 QVEVSN 1435
Q +VSN
Sbjct: 183 QAKVSN 200
>AW570005
Length = 413
Score = 85.5 bits (210), Expect = 2e-16
Identities = 50/141 (35%), Positives = 76/141 (53%)
Frame = -2
Query: 981 PPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTNLLLKDADFTFDDSCLQAFCRLKEALI 1040
PP + + +R FL GFYRRFIK ++++ PL++LL KD+ F + AF LK +
Sbjct: 409 PPRTARSLRGFLRLTGFYRRFIKGYAAMAAPLSHLLTKDS-FVWSPEADVAFQALKNVVT 233
Query: 1041 TAPIIQPPDWNLPFEIMCDASDYAVGVVLGQRKDKKMHAIYYASKTLDGAQVNYATTEKE 1100
++ PD+ PF + DAS +G VL Q + H I + SK V +T E
Sbjct: 232 NTLVLALPDFTKPFTVETDASGSDMGAVLSQ----EGHPIAFFSKEFCPKLVRSSTYVHE 65
Query: 1101 LLAVVYAIDKFRQYLVGSKII 1121
L A+ + K+RQYL+G ++
Sbjct: 64 LAAITNVVKKWRQYLLGHHLV 2
>AW184779
Length = 432
Score = 55.5 bits (132), Expect(2) = 1e-14
Identities = 23/69 (33%), Positives = 37/69 (53%)
Frame = +1
Query: 1241 LFRRGSDGIFRRCIPESEVSSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHL 1300
L++R D + RC+ E +L H S+G HA+ + KIL G++W ++ D +
Sbjct: 16 LYKRNHDMVLLRCVDAREAEQMLVEVHEGSFGTHANIHAMAQKILRVGYYWLTMESDCCI 195
Query: 1301 FISKCDKCQ 1309
+ KC KCQ
Sbjct: 196 HVWKCHKCQ 222
Score = 43.9 bits (102), Expect(2) = 1e-14
Identities = 20/47 (42%), Positives = 32/47 (67%), Gaps = 2/47 (4%)
Frame = +3
Query: 1333 VWGVDFMGPFP--SSFGNQYILVAVDYVSKWVEAIASPTNDAQVVIK 1377
+WG+D +G +S G+ +ILVA+DY +KWVEA++ + VVI+
Sbjct: 291 MWGIDVIGAIEPKASNGHHFILVAIDYFTKWVEAVSYASVTRSVVIR 431
>BE660092 weakly similar to GP|9884624|dbj retroelement pol polyprotein-like
{Arabidopsis thaliana}, partial (13%)
Length = 378
Score = 71.2 bits (173), Expect = 3e-12
Identities = 32/46 (69%), Positives = 38/46 (82%)
Frame = -3
Query: 1409 LLQKLGVRHKVATPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDWS 1454
LL+K GV H+V+TPYHPQT+GQ E+SNR+IK ILEK V SR DWS
Sbjct: 367 LLKKYGVVHRVSTPYHPQTNGQAEISNREIKRILEKIVQPSRKDWS 230
>BQ627806
Length = 435
Score = 46.2 bits (108), Expect(2) = 1e-11
Identities = 22/67 (32%), Positives = 39/67 (57%)
Frame = +1
Query: 1095 ATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDL 1154
+T +EL A+ A+ K+RQYL+G ++ TDH ++K L+++ P ++ L FD
Sbjct: 229 STYVRELAAITVAVKKWRQYLLGHHFVILTDHRSLKELMSQAVQTPEQQIYLARLMGFDY 408
Query: 1155 EIKDKKG 1161
I+ + G
Sbjct: 409 TIQYRAG 429
Score = 43.1 bits (100), Expect(2) = 1e-11
Identities = 25/67 (37%), Positives = 37/67 (54%)
Frame = +3
Query: 1015 LLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGVVLGQRKD 1074
LL+KD F +++ +AF +LK AL AP++ PD+N F + DAS +G +L Q
Sbjct: 3 LLVKD-QFHWNEEADRAFSQLKLALCQAPVLGLPDFNSSFVVETDASGIGMGAILSQNHH 179
Query: 1075 KKMHAIY 1081
IY
Sbjct: 180 PLAFFIY 200
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 65,831,031
Number of Sequences: 63676
Number of extensions: 959826
Number of successful extensions: 4816
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 4725
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4797
length of query: 1454
length of database: 12,639,632
effective HSP length: 109
effective length of query: 1345
effective length of database: 5,698,948
effective search space: 7665085060
effective search space used: 7665085060
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC135959.8


