

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
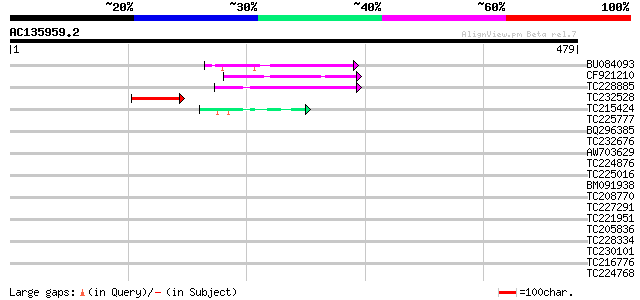
Query= AC135959.2 - phase: 0
(479 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BU084093 64 1e-10
CF921210 50 2e-06
TC228885 50 2e-06
TC232528 weakly similar to UP|Q6WAY5 (Q6WAY5) Gag/pol polyprotei... 48 8e-06
TC215424 similar to UP|Q9MA04 (Q9MA04) F20B17.16, partial (26%) 42 5e-04
TC225777 weakly similar to UP|Q868B4 (Q868B4) C. elegans GRL-25 ... 41 0.001
BQ296385 39 0.005
TC232676 similar to UP|Q6K709 (Q6K709) DNA-binding protein-like,... 39 0.007
AW703629 37 0.019
TC224876 weakly similar to UP|Q39835 (Q39835) Extensin, partial ... 36 0.043
TC225016 similar to UP|Q39835 (Q39835) Extensin, partial (32%) 36 0.043
BM091938 36 0.043
TC208770 weakly similar to UP|Q84ME1 (Q84ME1) At5g19080/T16G12_1... 35 0.057
TC227291 weakly similar to UP|Q9SFY4 (Q9SFY4) T22C5.20, partial ... 35 0.057
TC221951 UP|NO75_SOYBN (P08297) Early nodulin 75 precursor (N-75... 35 0.057
TC205836 32 0.066
TC228334 similar to UP|Q9SW80 (Q9SW80) Bel1-like homeodomain 2 (... 35 0.074
TC230101 similar to UP|Q41707 (Q41707) Extensin class 1 protein ... 35 0.074
TC216776 similar to UP|Q75WV0 (Q75WV0) ALG2-interacting protein ... 35 0.074
TC224768 similar to UP|Q39835 (Q39835) Extensin, partial (33%) 35 0.096
>BU084093
Length = 421
Score = 63.9 bits (154), Expect = 1e-10
Identities = 45/146 (30%), Positives = 63/146 (42%), Gaps = 16/146 (10%)
Frame = -1
Query: 165 SYPYAQYSQHPFFP----------PFYHQYPLPSGQPQVPVNAVVQQMQQQ------PPA 208
++ YAQ+ HP F P + P PQVP + + PP
Sbjct: 412 THQYAQH--HPSFSAHTGNASSSTPVQPKAPAQREAPQVPTPNTTRPVGNSNTTRNFPPR 239
Query: 209 QQQQHQQARPTFPPIPMLYAELLPTLLLRGHYTTRQGKPPPDPLPPRFRSDLKCDFHQGA 268
Q+ F P+PM Y +LLP+L+ G+ P P + + C +H G
Sbjct: 238 PFQE-------FTPLPMTYEDLLPSLIANHLAVVTPGRVLQPPFPKWYDPNATCKYHGGV 80
Query: 269 LGHDVEGCYALKYIVKKLINQGKLTF 294
GH VE C ALKY V+ L++ G LTF
Sbjct: 79 PGHSVEKCLALKYKVQHLMDAGWLTF 2
>CF921210
Length = 790
Score = 50.4 bits (119), Expect = 2e-06
Identities = 40/117 (34%), Positives = 54/117 (45%)
Frame = +2
Query: 181 YHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPPIPMLYAELLPTLLLRGHY 240
YH+ LP Q + P A + Q+ ++ + F PIP+ YA+LL LL
Sbjct: 236 YHKVCLPHFQ*RTPPLAQTKNTNQEVNFAARKPVE----FTPIPVSYADLLSYLLDNSMV 403
Query: 241 TTRQGKPPPDPLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLINQGKLTFENN 297
K PL + S+ C GAL H +E C ALK V+ LI+ G L FE N
Sbjct: 404 AITLAKVHQPPLF*GYDSNATCG---GALRHSIEHCRALKRKVQGLIDAGWLKFEEN 565
>TC228885
Length = 901
Score = 50.1 bits (118), Expect = 2e-06
Identities = 41/125 (32%), Positives = 53/125 (41%), Gaps = 1/125 (0%)
Frame = -3
Query: 174 HPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPT-FPPIPMLYAELLP 232
H P H+ LP Q + P A Q + Q+ +P F PIP+ YA+LLP
Sbjct: 743 HKGHP*INHKVCLPHFQ*RTPPLA-----QTKNTNQEMNFAARKPVEFTPIPVSYADLLP 579
Query: 233 TLLLRGHYTTRQGKPPPDPLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLINQGKL 292
LL K P + S+ C H A G +E ALK V+ LI+ G L
Sbjct: 578 YLLDNSMVAITLAKVHQPPFLREYDSNAMCACHGEAPGRSIEHYRALKRKVQGLIDAGWL 399
Query: 293 TFENN 297
FE N
Sbjct: 398 KFEEN 384
>TC232528 weakly similar to UP|Q6WAY5 (Q6WAY5) Gag/pol polyprotein
(Fragment), partial (3%)
Length = 449
Score = 48.1 bits (113), Expect = 8e-06
Identities = 22/44 (50%), Positives = 32/44 (72%)
Frame = +1
Query: 104 ATLDQLRYRGTPLGRSTLLVKAFHGTRKNVLGEIDLPITIDPET 147
+TLD+L + + L S+++V+AF GTR+ V GEIDLP+ I P T
Sbjct: 316 STLDKLPFNASHLKPSSMVVRAFDGTRREVRGEIDLPVQIGPHT 447
>TC215424 similar to UP|Q9MA04 (Q9MA04) F20B17.16, partial (26%)
Length = 1030
Score = 42.4 bits (98), Expect = 5e-04
Identities = 33/98 (33%), Positives = 38/98 (38%), Gaps = 4/98 (4%)
Frame = +3
Query: 161 QLPPSYPYAQYSQH--PFFPPFYHQ--YPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQA 216
Q+PP+ Y Q QH P+ PP H YP P P P A QPP
Sbjct: 48 QIPPNSNYHQNQQHHVPYAPPNPHHPHYPYPPPPPPPPPEA-----SYQPP--------- 185
Query: 217 RPTFPPIPMLYAELLPTLLLRGHYTTRQGKPPPDPLPP 254
P PP PM Y + + Q PPP PL P
Sbjct: 186 -PPPPPAPMYYPS--------NNQYSNQPPPPPPPLSP 272
>TC225777 weakly similar to UP|Q868B4 (Q868B4) C. elegans GRL-25 protein
(Corresponding sequence ZK643.8), partial (7%)
Length = 762
Score = 40.8 bits (94), Expect = 0.001
Identities = 29/93 (31%), Positives = 37/93 (39%), Gaps = 8/93 (8%)
Frame = +3
Query: 182 HQYPLPSGQPQVPVNAVVQQMQ-QQPPAQQQQHQQARPTFPPIPMLYAELLPTLLLRGHY 240
HQ LP QP V Q Q QQPP Q Q A PP P ++ + P L +
Sbjct: 132 HQVELPPPQPPVQQQQQPPQQQLQQPPQQPPQQHLAPQQLPPQPPIFQQTYPPFHLPQFH 311
Query: 241 -------TTRQGKPPPDPLPPRFRSDLKCDFHQ 266
+Q + PP P P F+ + HQ
Sbjct: 312 HHQPSLQFQQQQQQPPQPPPQLFQHQAQPPSHQ 410
>BQ296385
Length = 399
Score = 38.9 bits (89), Expect = 0.005
Identities = 32/98 (32%), Positives = 35/98 (35%), Gaps = 2/98 (2%)
Frame = +2
Query: 163 PPSYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPP 222
PP P Q QHP PP H + P P +V Q Q P RP PP
Sbjct: 77 PPRSP--QIPQHPILPPQIHSHSGPKPTKTNPGPSVGLQAQPSPLPPHPHRGHRRPLLPP 250
Query: 223 IPMLYAELLPTLLLRGHYTTRQGKPPP--DPLPPRFRS 258
P LR H G PP PLPP R+
Sbjct: 251 FP-----------LRPH-----GPPPQLRGPLPPPRRA 316
>TC232676 similar to UP|Q6K709 (Q6K709) DNA-binding protein-like, partial
(57%)
Length = 926
Score = 38.5 bits (88), Expect = 0.007
Identities = 29/83 (34%), Positives = 36/83 (42%), Gaps = 3/83 (3%)
Frame = +1
Query: 181 YHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPPIPMLYAELLPTLLLRGH- 239
Y + PL Q + + QQ QQQ QQQQ QQ++ + M L P LL G
Sbjct: 451 YERLPLEDDQGEEEMQVQQQQQQQQQQQQQQQQQQSQGLGEQVSMPMYNLPPNLLHNGQN 630
Query: 240 --YTTRQGKPPPDPLPPRFRSDL 260
+ G PP PP F S L
Sbjct: 631 MPHDVFWGAPPRP--PPSF*SPL 693
>AW703629
Length = 421
Score = 37.0 bits (84), Expect = 0.019
Identities = 21/63 (33%), Positives = 24/63 (37%)
Frame = +1
Query: 163 PPSYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPP 222
PP P Q QHP PP H + P P +V Q Q P RP PP
Sbjct: 97 PPRSP--QIPQHPILPPQIHSHSGPKPTKTNPGPSVGLQAQPSPLPPHPHRGHRRPLLPP 270
Query: 223 IPM 225
P+
Sbjct: 271 FPL 279
>TC224876 weakly similar to UP|Q39835 (Q39835) Extensin, partial (66%)
Length = 1340
Score = 35.8 bits (81), Expect = 0.043
Identities = 27/92 (29%), Positives = 36/92 (38%)
Frame = +3
Query: 163 PPSYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPP 222
PP +PY HP+ P H +P P P P V PP + ++ P+ PP
Sbjct: 834 PPPHPYPHPHPHPYPHPHPHPHPHPHPHPH-PHPPYVYPSPPPPPKKPYKY----PSPPP 998
Query: 223 IPMLYAELLPTLLLRGHYTTRQGKPPPDPLPP 254
Y P + HY+ PPP P P
Sbjct: 999 PVHHYPHPHPPFV---HYS-----PPPSPKKP 1070
Score = 28.9 bits (63), Expect = 5.3
Identities = 27/97 (27%), Positives = 34/97 (34%), Gaps = 4/97 (4%)
Frame = +1
Query: 163 PPSYPYAQYSQHPFFPP----FYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARP 218
PP Y Y PP +YH P PS P P PP +++ + P
Sbjct: 214 PPKYYYHSPPPPSPSPPKKPYYYHSPPPPSPTP--PKKPYYYHSPPPPPPKKKPYYYHSP 387
Query: 219 TFPPIPMLYAELLPTLLLRGHYTTRQGKPPPDPLPPR 255
PP P P +Y PPP P PP+
Sbjct: 388 P-PPSP-------PPPKKPYYY---HSPPPPSPSPPK 465
>TC225016 similar to UP|Q39835 (Q39835) Extensin, partial (32%)
Length = 973
Score = 35.8 bits (81), Expect = 0.043
Identities = 27/92 (29%), Positives = 36/92 (38%)
Frame = +2
Query: 163 PPSYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPP 222
PP +PY HP+ P H +P P P P V PP + ++ P+ PP
Sbjct: 242 PPPHPYPHPHPHPYPHPHPHPHPHPHPHPH-PHPPYVYPSPPPPPKKPYKY----PSPPP 406
Query: 223 IPMLYAELLPTLLLRGHYTTRQGKPPPDPLPP 254
Y P + HY+ PPP P P
Sbjct: 407 PVHHYPHPHPPFV---HYS-----PPPSPKKP 478
Score = 30.0 bits (66), Expect = 2.4
Identities = 24/90 (26%), Positives = 29/90 (31%)
Frame = +2
Query: 164 PSYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPPI 223
P Y Y P P +YH P PS P P PP ++ + + P PP
Sbjct: 41 PPYVYPSPPPPPKKPYYYHSPPPPSPPPPKPY--YYHSPPPPPPKKKPYYYHSPPPPPPK 214
Query: 224 PMLYAELLPTLLLRGHYTTRQGKPPPDPLP 253
Y P PPP P P
Sbjct: 215 KKPYYYHSP--------------PPPHPYP 262
>BM091938
Length = 421
Score = 35.8 bits (81), Expect = 0.043
Identities = 27/92 (29%), Positives = 36/92 (38%)
Frame = +2
Query: 163 PPSYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPP 222
PP +PY HP+ P H +P P P P V PP + ++ P+ PP
Sbjct: 116 PPPHPYPHPHPHPYPHPHPHPHPHPHPHPH-PHPPYVYPSPPPPPKKPYKY----PSPPP 280
Query: 223 IPMLYAELLPTLLLRGHYTTRQGKPPPDPLPP 254
Y P + HY+ PPP P P
Sbjct: 281 PVHHYPHPHPPFV---HYS-----PPPSPKKP 352
>TC208770 weakly similar to UP|Q84ME1 (Q84ME1) At5g19080/T16G12_120, partial
(30%)
Length = 753
Score = 35.4 bits (80), Expect = 0.057
Identities = 30/89 (33%), Positives = 31/89 (34%)
Frame = +1
Query: 163 PPSYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPP 222
PPSY Y S HP PP Y LPS P P PP Q +
Sbjct: 40 PPSYYYHTES-HPAPPPQPQGYFLPSATPYAP-------PPPMPPPPPPQTHSISFYYSS 195
Query: 223 IPMLYAELLPTLLLRGHYTTRQGKPPPDP 251
P Y PTL R HY P P P
Sbjct: 196 NPNSYG--TPTLAPRFHYPQHYYHPQPQP 276
>TC227291 weakly similar to UP|Q9SFY4 (Q9SFY4) T22C5.20, partial (7%)
Length = 1393
Score = 35.4 bits (80), Expect = 0.057
Identities = 26/96 (27%), Positives = 40/96 (41%)
Frame = +1
Query: 167 PYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPPIPML 226
P AQ P P Y+Q QVP + + + PP Q + P+ PP+P +
Sbjct: 439 PLAQIQGTPMHP--YNQ--------QVPPSIMPPPLSSLPPPQPEMPPPLPPSPPPLPQV 588
Query: 227 YAELLPTLLLRGHYTTRQGKPPPDPLPPRFRSDLKC 262
L+P L + PPP LP + +++C
Sbjct: 589 QPPLVPPL------PSSPPPPPPPQLPVQEPVNMEC 678
>TC221951 UP|NO75_SOYBN (P08297) Early nodulin 75 precursor (N-75) (NGM-75),
complete
Length = 1179
Score = 35.4 bits (80), Expect = 0.057
Identities = 25/98 (25%), Positives = 42/98 (42%), Gaps = 4/98 (4%)
Frame = +1
Query: 163 PPSYPYAQYSQHPFFPPFYHQYPLPSGQP---QVPVNAVVQQMQQQPPAQQQQHQQARPT 219
PP+Y + + P++PP H +P P QP + P + ++ PP H++ P
Sbjct: 121 PPTYEPPPFYKPPYYPPPVH-HPPPEYQPPHEKTPPEYLPPPHEKPPPEYLPPHEKPPPE 297
Query: 220 F-PPIPMLYAELLPTLLLRGHYTTRQGKPPPDPLPPRF 256
+ PP E P H + +PP + PP +
Sbjct: 298 YQPPHEKPPHENPPPEHQPPHEKPPEHQPPHEKPPPEY 411
Score = 32.3 bits (72), Expect = 0.48
Identities = 29/104 (27%), Positives = 37/104 (34%), Gaps = 10/104 (9%)
Frame = +1
Query: 161 QLPPSYPYAQYSQHPFFPPFY---HQYPLPSGQP-------QVPVNAVVQQMQQQPPAQQ 210
Q P P H PP Y H+ P P QP + P ++ PP Q
Sbjct: 544 QPPHEKPPEHQPPHEKPPPEYQPPHEKPPPEYQPPQEKPPHEKPPPEYQPPHEKPPPEHQ 723
Query: 211 QQHQQARPTFPPIPMLYAELLPTLLLRGHYTTRQGKPPPDPLPP 254
H++ P +PP Y + P Y KPPP PP
Sbjct: 724 PPHEKPPPVYPP---PYEKPPPV------YEPPYEKPPPVVYPP 828
>TC205836
Length = 1114
Score = 32.0 bits (71), Expect(2) = 0.066
Identities = 15/38 (39%), Positives = 17/38 (44%)
Frame = +3
Query: 218 PTFPPIPMLYAELLPTLLLRGHYTTRQGKPPPDPLPPR 255
PT P P + T+ TR KPPPDP P R
Sbjct: 270 PTTNPTPTSTTTSISTMTTTTTICTRNSKPPPDPTPTR 383
Score = 21.9 bits (45), Expect(2) = 0.066
Identities = 13/33 (39%), Positives = 16/33 (48%)
Frame = +1
Query: 182 HQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQ 214
H+ P P PQ P+ PP QQ+QHQ
Sbjct: 157 HRNPTP---PQPPL----------PPPQQRQHQ 216
>TC228334 similar to UP|Q9SW80 (Q9SW80) Bel1-like homeodomain 2
(AT4g36870/C7A10_490), partial (9%)
Length = 910
Score = 35.0 bits (79), Expect = 0.074
Identities = 22/66 (33%), Positives = 35/66 (52%), Gaps = 3/66 (4%)
Frame = +2
Query: 192 QVPVNAVVQQMQQQPPAQQQQHQQAR---PTFPPIPMLYAELLPTLLLRGHYTTRQGKPP 248
Q P + Q+ QQQQ++QA+ P+ PPI L++ +L ++ + T+Q PP
Sbjct: 140 QRPQHQQQQRHHHHHQRQQQQNRQAKDPMPSMPPIATLHS-MLQ*IIDKASRKTKQRSPP 316
Query: 249 PDPLPP 254
P PP
Sbjct: 317 HPPPPP 334
>TC230101 similar to UP|Q41707 (Q41707) Extensin class 1 protein precursor,
partial (47%)
Length = 944
Score = 35.0 bits (79), Expect = 0.074
Identities = 27/94 (28%), Positives = 36/94 (37%), Gaps = 3/94 (3%)
Frame = +2
Query: 159 AAQLPPSYPYAQY---SQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQ 215
+A PP+Y Y S P P +Y P PS P P ++PPA + +
Sbjct: 554 SASPPPAYYYKSPPPPSPSPTPPYYYKTAPPPSPSPPPPYYYKSPPPPEKPPAPTPYYNK 733
Query: 216 ARPTFPPIPMLYAELLPTLLLRGHYTTRQGKPPP 249
+ P PP P Y P Y + PPP
Sbjct: 734 SPP--PPSPTPYYYKSPPTPSPTPYYYKSAPPPP 829
>TC216776 similar to UP|Q75WV0 (Q75WV0) ALG2-interacting protein X, partial
(11%)
Length = 855
Score = 35.0 bits (79), Expect = 0.074
Identities = 32/105 (30%), Positives = 39/105 (36%), Gaps = 10/105 (9%)
Frame = +3
Query: 164 PSYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPAQQQQHQQARPTFPPI 223
P PY +QH PP YH P S P P Q+ QQPPA H+ +P +P
Sbjct: 261 PPPPYGGPAQHHQPPPPYH-IPPSSTAPYPP-----PQVHQQPPA---NHEYGQPAYPG- 410
Query: 224 PMLYAELLPTLLLRGHYTTRQGKPP----------PDPLPPRFRS 258
RG Y Q + P P P PP +S
Sbjct: 411 ------------WRGPYYNAQAQQPGSVPRPPYTIPSPYPPPHQS 509
>TC224768 similar to UP|Q39835 (Q39835) Extensin, partial (33%)
Length = 483
Score = 34.7 bits (78), Expect = 0.096
Identities = 27/100 (27%), Positives = 36/100 (36%), Gaps = 8/100 (8%)
Frame = +3
Query: 163 PPSYPYAQYSQHPFFPPFYHQYPLPSGQP---QVPVNAVVQQMQQQPPAQQQQHQQARPT 219
PP PY +S P P+Y+ P P P + P PP ++ + + P
Sbjct: 174 PPKKPYYYHSPPPPKKPYYYHSPPPPSPPPPYKKPYYYHSPPPPSPPPPKKPYYYHSPPP 353
Query: 220 FPPIPMLY-----AELLPTLLLRGHYTTRQGKPPPDPLPP 254
P P Y P HY + PPP P PP
Sbjct: 354 PPKKPYYYHSPPPPSPPPPYKKPYHYVS---PPPPSPSPP 464
Score = 29.6 bits (65), Expect = 3.1
Identities = 23/109 (21%), Positives = 37/109 (33%)
Frame = +3
Query: 147 TFLITFQVMDINAAQLPPSYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQP 206
T + ++ ++ ++ Y S P P+Y+ P P P P P
Sbjct: 39 TLTLVLAIVSLSLPSQISAHNYLYSSPPPPKKPYYYHSPPPPSPP--PPKKPYYYHSPPP 212
Query: 207 PAQQQQHQQARPTFPPIPMLYAELLPTLLLRGHYTTRQGKPPPDPLPPR 255
P + + P PP P + +Y PPP P PP+
Sbjct: 213 PKKPYYYHSPPPPSPPPPYK----------KPYYY--HSPPPPSPPPPK 323
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,223,512
Number of Sequences: 63676
Number of extensions: 388976
Number of successful extensions: 3556
Number of sequences better than 10.0: 150
Number of HSP's better than 10.0 without gapping: 3107
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3397
length of query: 479
length of database: 12,639,632
effective HSP length: 101
effective length of query: 378
effective length of database: 6,208,356
effective search space: 2346758568
effective search space used: 2346758568
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC135959.2


