

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
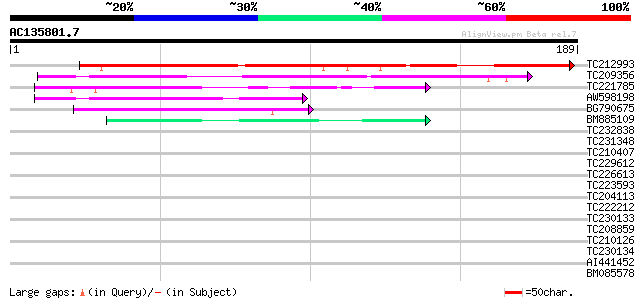
Query= AC135801.7 - phase: 0
(189 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC212993 weakly similar to UP|Q9Y1M2 (Q9Y1M2) P68 RNA helicase, ... 182 8e-47
TC209356 similar to UP|Q7Q0C7 (Q7Q0C7) AgCP9374 (Fragment), part... 55 2e-08
TC221785 similar to UP|Q6NNJ2 (Q6NNJ2) At5g01790, partial (21%) 54 6e-08
AW598198 48 2e-06
BG790675 44 4e-05
BM885109 44 4e-05
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 38 0.003
TC231348 homologue to UP|Q6NNJ2 (Q6NNJ2) At5g01790, partial (6%) 36 0.009
TC210407 similar to UP|O49656 (O49656) Predicted protein, partia... 35 0.021
TC229612 35 0.027
TC226613 similar to UP|Q8H0V8 (Q8H0V8) Spliceosome associated pr... 33 0.060
TC223593 similar to GB|AAN28762.1|23505805|AY143823 At1g71970/F1... 33 0.10
TC204113 homologue to UP|Q9LD28 (Q9LD28) Histone H2A, partial (96%) 33 0.10
TC222212 similar to GB|AAO64844.1|29028930|BT005909 At3g53320 {A... 32 0.13
TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 32 0.13
TC208859 32 0.18
TC210126 similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C2... 32 0.18
TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Pro... 32 0.18
AI441452 32 0.23
BM085578 similar to GP|9757868|dbj| formin-like protein {Arabido... 31 0.30
>TC212993 weakly similar to UP|Q9Y1M2 (Q9Y1M2) P68 RNA helicase, partial (4%)
Length = 625
Score = 182 bits (462), Expect = 8e-47
Identities = 106/173 (61%), Positives = 123/173 (70%), Gaps = 8/173 (4%)
Frame = +3
Query: 24 ISYYRS-GEGVPFKWEMQPGIAKESSPKEVLPPLTPPPKNLSLGLPKPSILELKKPASTM 82
+SYYRS GEGVPFKWEMQPGIAKE P+E LPPL+PPP LSLGLPKP I + K ST
Sbjct: 3 VSYYRSSGEGVPFKWEMQPGIAKEQQPREDLPPLSPPPALLSLGLPKPCIPDTK--PSTR 176
Query: 83 SKLKFWKKRVVKIKSKKPQEV---CFHEDFDI--LSRLDCSSDSE--SMVSPRGSSFSSS 135
S+L+FWKKRV + KSKK CFHED D+ L LDC+SDSE SM SPRGSSF SS
Sbjct: 177 SRLRFWKKRVKRGKSKKSSSSSHDCFHEDDDVIGLDILDCNSDSESNSMASPRGSSF-SS 353
Query: 136 SSSMSLMKSTRSSLHSVCSSCSEGNTKQVSRKPSTLGCFPMHISRVLVSITRR 188
SSMS MKS +++ ++V R+PSTLGCFPMH++RV VSI RR
Sbjct: 354 WSSMSFMKSPTTTI------------RKVYRRPSTLGCFPMHVTRVFVSIARR 476
>TC209356 similar to UP|Q7Q0C7 (Q7Q0C7) AgCP9374 (Fragment), partial (3%)
Length = 934
Score = 55.1 bits (131), Expect = 2e-08
Identities = 50/169 (29%), Positives = 77/169 (44%), Gaps = 4/169 (2%)
Frame = +1
Query: 10 KILSRRSSVGCSSRISYYRSGEGVPFKWEMQPGIAKESSPKEVLPPLTPPPKNLSLGLPK 69
++LS+ SS+ S +R VPF WE +PG K ++ LPPLTPP
Sbjct: 166 RLLSKESSMANPS----FRIALAVPFVWESEPGTPKHRFSEDTLPPLTPP---------- 303
Query: 70 PSILELKKPASTMSKLKFWKKRVVKIKSKKPQEVCFHEDFDILSRLDCSSDSESMVSPRG 129
P+ S + + K+ K SK + +++ ++ SS S S S
Sbjct: 304 --------PSYHFSTINSY*KKEKKHSSKSSLLLTMLPKLNLIKKMILSS-SSSYPSSWP 456
Query: 130 SSFSSSSSSMSLMKSTRSSLHSVCSSCSE--GNTKQV--SRKPSTLGCF 174
SS SSSS + + K + L S SSC++ G+ + V + PS+ CF
Sbjct: 457 SSSSSSSKVVPMAKFGKKKLLSYGSSCNDFKGDCEGVGGASSPSSQLCF 603
>TC221785 similar to UP|Q6NNJ2 (Q6NNJ2) At5g01790, partial (21%)
Length = 686
Score = 53.5 bits (127), Expect = 6e-08
Identities = 47/134 (35%), Positives = 63/134 (46%), Gaps = 2/134 (1%)
Frame = +3
Query: 9 NKILSRRSSVG-CSSRISYY-RSGEGVPFKWEMQPGIAKESSPKEVLPPLTPPPKNLSLG 66
++++S+ +S+ SSR+ YY + VPF WE QPG K S + LPPLTPPP S
Sbjct: 123 SRLMSKETSMANSSSRVFYYGETSIAVPFMWEAQPGTPKHPSSETCLPPLTPPPSYYS-- 296
Query: 67 LPKPSILELKKPASTMSKLKFWKKRVVKIKSKKPQEVCFHEDFDILSRLDCSSDSESMVS 126
++ SK K + SK CF F I++R + VS
Sbjct: 297 -------------NSKSKNK-------RRNSKSNIFSCFMPKF-IVAR-------KGHVS 392
Query: 127 PRGSSFSSSSSSMS 140
P S SSSSSS S
Sbjct: 393 PTSSRSSSSSSSSS 434
>AW598198
Length = 410
Score = 48.1 bits (113), Expect = 2e-06
Identities = 30/91 (32%), Positives = 44/91 (47%)
Frame = +3
Query: 9 NKILSRRSSVGCSSRISYYRSGEGVPFKWEMQPGIAKESSPKEVLPPLTPPPKNLSLGLP 68
+++LS+ SS+ S +R VPF WE QPG K + ++ LPPLTPPP +
Sbjct: 123 SRLLSKESSMSKPS----FRMAVAVPFVWESQPGTPKYTFSEDTLPPLTPPPSYFVNFIT 290
Query: 69 KPSILELKKPASTMSKLKFWKKRVVKIKSKK 99
K + KP S+ + K+ KK
Sbjct: 291 KKN-----KPLKKRSRSNLFLALFPKLNLKK 368
>BG790675
Length = 417
Score = 43.9 bits (102), Expect = 4e-05
Identities = 26/81 (32%), Positives = 35/81 (43%), Gaps = 1/81 (1%)
Frame = +1
Query: 22 SRISYYRSGEGVPFKWEMQPGIAKESSPKEVLPPLTPPPKNLSLGLPKPSILELKKPAST 81
S +Y+ +PF WE QPG K + LPPLTPPP K + K +
Sbjct: 175 SNEAYHVESSSIPFVWESQPGTPKVRFKENSLPPLTPPPSYFQNATKKATTKAKNKNSPK 354
Query: 82 MSKLK-FWKKRVVKIKSKKPQ 101
S L+ + KR + PQ
Sbjct: 355 SSFLQTLFPKRATRKGGVPPQ 417
>BM885109
Length = 421
Score = 43.9 bits (102), Expect = 4e-05
Identities = 36/108 (33%), Positives = 43/108 (39%)
Frame = +1
Query: 33 VPFKWEMQPGIAKESSPKEVLPPLTPPPKNLSLGLPKPSILELKKPASTMSKLKFWKKRV 92
VPF WE QPG K + LPPLTPPP S S +K + + K
Sbjct: 16 VPFTWEAQPGTPKHPLSETSLPPLTPPPSYFS------------NKKSHNAKSRNYSKAS 159
Query: 93 VKIKSKKPQEVCFHEDFDILSRLDCSSDSESMVSPRGSSFSSSSSSMS 140
S P+ V S + VSP SS+S SSSS S
Sbjct: 160 NIFSSILPRRV--------------GSRNSHPVSPASSSWSLSSSSSS 261
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 37.7 bits (86), Expect = 0.003
Identities = 24/44 (54%), Positives = 25/44 (56%)
Frame = -3
Query: 118 SSDSESMVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNT 161
SS S S+ SP SS SSSSSS S ST SS S SS S T
Sbjct: 402 SSSSSSLSSPSSSSSSSSSSSSSSASSTTSSTDSYSSSSSSTKT 271
Score = 29.3 bits (64), Expect = 1.1
Identities = 21/53 (39%), Positives = 25/53 (46%)
Frame = -3
Query: 118 SSDSESMVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTKQVSRKPST 170
SS S S S S SSSSSS+S S+ SS S SS + T S+
Sbjct: 447 SSSSSSSSSSPPSGASSSSSSLSSPSSSSSSSSSSSSSSASSTTSSTDSYSSS 289
Score = 28.9 bits (63), Expect = 1.5
Identities = 31/93 (33%), Positives = 36/93 (38%), Gaps = 11/93 (11%)
Frame = -3
Query: 89 KKRVVKIKSKKPQEVCFHEDFDILSRLDCSSDSESM----------VSPRGSSFSSSSSS 138
K R+ K C S S SES+ SP SS SSSSSS
Sbjct: 597 KGRIRYSVPKSSSSPCSSSSSSSSSSSSSESSSESLSSSSGSPPRWTSPSSSSSSSSSSS 418
Query: 139 -MSLMKSTRSSLHSVCSSCSEGNTKQVSRKPST 170
S S+ SSL S SS S ++ S ST
Sbjct: 417 PPSGASSSSSSLSSPSSSSSSSSSSSSSSASST 319
>TC231348 homologue to UP|Q6NNJ2 (Q6NNJ2) At5g01790, partial (6%)
Length = 591
Score = 36.2 bits (82), Expect = 0.009
Identities = 41/137 (29%), Positives = 52/137 (37%), Gaps = 30/137 (21%)
Frame = +2
Query: 38 EMQPGIAKESSPKEVLPPLTPPP------------------KNLSLGL-PKPSILELKK- 77
E QPG K + ++ LPPLTPPP N+ L L PK L LKK
Sbjct: 5 EAQPGTPKYTFSEDTLPPLTPPPSYFVNSNKKNKPLKKRSRSNIFLALFPK---LNLKKK 175
Query: 78 ----PASTMSKLKFW------KKRVVKIKSKKPQEVCFHEDFDILSRLDCSSDSESMVSP 127
P+S S W K V K + + + F FD + S S S SP
Sbjct: 176 NILSPSSPTSPSSLWSTSDHSSKVVPMEKHGRRRFLSFGSSFDFRGDEEEESASASATSP 355
Query: 128 RGSSFSSSSSSMSLMKS 144
+ S S S + S
Sbjct: 356 TSTLCFGISRSTSTLAS 406
>TC210407 similar to UP|O49656 (O49656) Predicted protein, partial (33%)
Length = 1010
Score = 35.0 bits (79), Expect = 0.021
Identities = 35/121 (28%), Positives = 53/121 (42%), Gaps = 4/121 (3%)
Frame = +3
Query: 68 PKPSILELK-KPASTMSKLKFWKKRVVKIKSKKPQEVCFHEDFDILSRLD---CSSDSES 123
P P+IL + + + T+ + W++ S++P I S CSS S S
Sbjct: 141 PPPAILGFR*RCSPTLPSRRPWRRWAATSPSRRPSGSTTPPPSPITSSTATTFCSSSSSS 320
Query: 124 MVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTKQVSRKPSTLGCFPMHISRVLV 183
SP SS S+SS+S S + TRS+ S C + + S S+ P + S L
Sbjct: 321 PSSPSPSSSSNSSASPSSLP-TRSNQKSACPWPKPSSATKTSCACSSSSSAPSNSSLTLP 497
Query: 184 S 184
S
Sbjct: 498 S 500
>TC229612
Length = 684
Score = 34.7 bits (78), Expect = 0.027
Identities = 38/139 (27%), Positives = 60/139 (42%), Gaps = 8/139 (5%)
Frame = +1
Query: 26 YYRSGEGVPFKWEMQPGIAKESSPKEVLPPLTPPPKNLSLGLPKPSILELKKPASTMSKL 85
+ R VPF WE++PGI K+ E P+TP L L P + E +KP +
Sbjct: 7 HVREPPSVPFIWEVKPGIPKKDWKPEAEIPITP----LKLIASVPFVWE-EKPGKPLPNF 171
Query: 86 KF-----WKKRVVKIKSKKPQEVCF---HEDFDILSRLDCSSDSESMVSPRGSSFSSSSS 137
K ++ + S V H+D + S SSD+ES+ + +FS
Sbjct: 172 SVDHPVPSKPLLIHVASSSGFSVACNFGHDDKEKGSLSLSSSDNESITTLDLEAFSFDED 351
Query: 138 SMSLMKSTRSSLHSVCSSC 156
+S SS+ S+ ++C
Sbjct: 352 -----ESFISSVPSLLANC 393
>TC226613 similar to UP|Q8H0V8 (Q8H0V8) Spliceosome associated protein-like
(At4g21660), partial (59%)
Length = 1844
Score = 33.5 bits (75), Expect = 0.060
Identities = 24/74 (32%), Positives = 34/74 (45%), Gaps = 5/74 (6%)
Frame = -1
Query: 4 FEIESNKILSRRSSVGCSSRISYYRSGEGVPFKWEMQPGIAKESSPKEVLPP-----LTP 58
F + S ++L + +S SS S R + P +WE + +V PP L P
Sbjct: 1076 FFLPSLQVLDKEASQAPSSVASGGR*HQVFPLQWECLTDYQQTEYQLQVPPPPFLLLLPP 897
Query: 59 PPKNLSLGLPKPSI 72
PP L L LP PS+
Sbjct: 896 PPPPLPLPLPLPSL 855
Score = 28.1 bits (61), Expect = 2.5
Identities = 19/41 (46%), Positives = 23/41 (55%)
Frame = -3
Query: 115 LDCSSDSESMVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSS 155
+ SS S S+ S SS SSSSSS S KS + + S SS
Sbjct: 945 IPASSSSSSISSSSSSSSSSSSSSSSSSKSPQCLVLSTGSS 823
>TC223593 similar to GB|AAN28762.1|23505805|AY143823 At1g71970/F17M19_12
{Arabidopsis thaliana;} , partial (16%)
Length = 421
Score = 32.7 bits (73), Expect = 0.10
Identities = 24/77 (31%), Positives = 32/77 (41%)
Frame = +2
Query: 32 GVPFKWEMQPGIAKESSPKEVLPPLTPPPKNLSLGLPKPSILELKKPASTMSKLKFWKKR 91
GVPF WE PGI K+ + K+ L + SL +L L P +T K
Sbjct: 212 GVPFSWEHLPGIPKKQNSKKKL-------QESSL-----KLLPLPPPTTTHCSSKKHSHE 355
Query: 92 VVKIKSKKPQEVCFHED 108
+IK K + F D
Sbjct: 356 ETRIKKKNSIQSVFQRD 406
>TC204113 homologue to UP|Q9LD28 (Q9LD28) Histone H2A, partial (96%)
Length = 803
Score = 32.7 bits (73), Expect = 0.10
Identities = 21/65 (32%), Positives = 31/65 (47%), Gaps = 2/65 (3%)
Frame = +3
Query: 42 GIAK--ESSPKEVLPPLTPPPKNLSLGLPKPSILELKKPASTMSKLKFWKKRVVKIKSKK 99
G AK +SP V P + P LS+ P P L P S +S+L+FW ++ +
Sbjct: 165 GAAKPASNSPSAVSPVSSRPENTLSVSAPAP--LSTSPPYSNISRLRFWNWLETLRETTR 338
Query: 100 PQEVC 104
E+C
Sbjct: 339 RLELC 353
>TC222212 similar to GB|AAO64844.1|29028930|BT005909 At3g53320 {Arabidopsis
thaliana;} , partial (5%)
Length = 686
Score = 32.3 bits (72), Expect = 0.13
Identities = 36/105 (34%), Positives = 46/105 (43%), Gaps = 4/105 (3%)
Frame = +1
Query: 67 LPKPSILELKKPA----STMSKLKFWKKRVVKIKSKKPQEVCFHEDFDILSRLDCSSDSE 122
+PKP+ L K P+ ST +K VVK S+ + LSRL ++
Sbjct: 58 VPKPT-LPSKSPSGPTVSTRTKSVTSTSSVVKTPSRVASRNKAEPEISSLSRLMSATKLS 234
Query: 123 SMVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTKQVSRK 167
S +SP SS S SSS S ST S VC+S SRK
Sbjct: 235 SSISP-ASSISDWSSSES--SSTTSMAKRVCNSSRPSIDSGSSRK 360
>TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 850
Score = 32.3 bits (72), Expect = 0.13
Identities = 25/63 (39%), Positives = 31/63 (48%)
Frame = -1
Query: 115 LDCSSDSESMVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTKQVSRKPSTLGCF 174
L S SES SP S SSSSS SL SS S S S ++ ++ S+ CF
Sbjct: 409 LASGSSSESASSPPSSPEKSSSSSSSL-----SSSPSFTLSSSSSSSVSLASLFSSEVCF 245
Query: 175 PMH 177
P+H
Sbjct: 244 PLH 236
Score = 27.3 bits (59), Expect = 4.3
Identities = 22/55 (40%), Positives = 27/55 (49%)
Frame = -1
Query: 118 SSDSESMVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTKQVSRKPSTLG 172
SS S SP + SSSSSS+SL S + S S+GN PST+G
Sbjct: 346 SSSSSLSSSPSFTLSSSSSSSVSLASLFSSEVCFPLHSWSKGNL------PSTIG 200
>TC208859
Length = 858
Score = 32.0 bits (71), Expect = 0.18
Identities = 18/48 (37%), Positives = 23/48 (47%)
Frame = +3
Query: 33 VPFKWEMQPGIAKESSPKEVLPPLTPPPKNLSLGLPKPSILELKKPAS 80
VPF+WE PG + P L P + PP L L P L + PA+
Sbjct: 141 VPFRWEQVPG---KPRPSTALVPFSNPPDLLPKCLELPPRLLIPSPAT 275
>TC210126 similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23), partial
(7%)
Length = 735
Score = 32.0 bits (71), Expect = 0.18
Identities = 22/58 (37%), Positives = 26/58 (43%)
Frame = -3
Query: 118 SSDSESMVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTKQVSRKPSTLGCFP 175
SS S S S SS SSSSSS+SL S+ S + S S G+ P P
Sbjct: 571 SSSSSSSSSSSSSSPSSSSSSLSLSSSSSSLPPPLASGSSSGSPSSSPSSPENSSSSP 398
>TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23),
partial (8%)
Length = 581
Score = 32.0 bits (71), Expect = 0.18
Identities = 24/63 (38%), Positives = 31/63 (49%)
Frame = -3
Query: 115 LDCSSDSESMVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTKQVSRKPSTLGCF 174
L S SES SP S SSSSS S+ S S SS S ++ ++ S+ CF
Sbjct: 408 LASGSSSESASSPPSSPEKSSSSS----SSSSPSSPSFTSSSSSSSSVSLASLFSSEVCF 241
Query: 175 PMH 177
P+H
Sbjct: 240 PLH 232
Score = 30.0 bits (66), Expect = 0.67
Identities = 21/48 (43%), Positives = 23/48 (47%)
Frame = -3
Query: 111 ILSRLDCSSDSESMVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSE 158
+ S SS S S SP SS SSSSSS S S+ S S SE
Sbjct: 528 VSSSSSSSSSSSSSSSPSSSSLSSSSSSSSSSSSSLSPPPLASGSSSE 385
Score = 30.0 bits (66), Expect = 0.67
Identities = 23/55 (41%), Positives = 28/55 (50%)
Frame = -3
Query: 118 SSDSESMVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTKQVSRKPSTLG 172
SS S S SP +S SSSSSS+SL S + S+GN PST+G
Sbjct: 342 SSSSSSPSSPSFTSSSSSSSSVSLASLFSSEVCFPLHGWSKGNL------PSTIG 196
Score = 27.7 bits (60), Expect = 3.3
Identities = 22/57 (38%), Positives = 29/57 (50%), Gaps = 4/57 (7%)
Frame = -3
Query: 118 SSDSESMVSPRGSSFSSSSSSMS----LMKSTRSSLHSVCSSCSEGNTKQVSRKPST 170
SS S S +S SS SSSSSS+S S+ S S SS + ++ S PS+
Sbjct: 486 SSPSSSSLSSSSSSSSSSSSSLSPPPLASGSSSESASSPPSSPEKSSSSSSSSSPSS 316
Score = 27.7 bits (60), Expect = 3.3
Identities = 19/43 (44%), Positives = 22/43 (50%)
Frame = -3
Query: 130 SSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTKQVSRKPSTLG 172
SS SSSSSS S S SS S SS S ++ +S P G
Sbjct: 525 SSSSSSSSSSSSSSSPSSSSLSSSSSSSSSSSSSLSPPPLASG 397
>AI441452
Length = 424
Score = 31.6 bits (70), Expect = 0.23
Identities = 15/43 (34%), Positives = 21/43 (47%)
Frame = +2
Query: 26 YYRSGEGVPFKWEMQPGIAKESSPKEVLPPLTPPPKNLSLGLP 68
+ R VPF WE++PGI K+ E P + P L +P
Sbjct: 17 HVREPPSVPFIWEVKPGIPKKDWKPEPEPEVPKTPLKLIASVP 145
>BM085578 similar to GP|9757868|dbj| formin-like protein {Arabidopsis
thaliana}, partial (6%)
Length = 424
Score = 31.2 bits (69), Expect = 0.30
Identities = 16/63 (25%), Positives = 26/63 (40%)
Frame = +3
Query: 36 KWEMQPGIAKESSPKEVLPPLTPPPKNLSLGLPKPSILELKKPASTMSKLKFWKKRVVKI 95
+WEM + P PPLTPP + L P + ++ P ++ + + K
Sbjct: 111 QWEMPSPLTPVDQPVSRPPPLTPPSRPFVLQTPNTKVSPVELPPASSQNFEEGSEETSKP 290
Query: 96 KSK 98
K K
Sbjct: 291 KLK 299
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.128 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,736,046
Number of Sequences: 63676
Number of extensions: 197289
Number of successful extensions: 2617
Number of sequences better than 10.0: 192
Number of HSP's better than 10.0 without gapping: 1989
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2320
length of query: 189
length of database: 12,639,632
effective HSP length: 92
effective length of query: 97
effective length of database: 6,781,440
effective search space: 657799680
effective search space used: 657799680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC135801.7


