

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135797.3 - phase: 0 /pseudo
(460 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
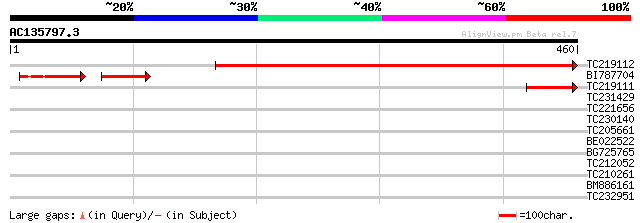
Score E
Sequences producing significant alignments: (bits) Value
TC219112 weakly similar to UP|Q94AA1 (Q94AA1) AT3g09470/F11F8_4,... 437 e-123
BI787704 similar to GP|15146218|gb| AT3g09470/F11F8_4 {Arabidops... 67 5e-20
TC219111 61 1e-09
TC231429 weakly similar to GB|AAP68235.1|31711758|BT008796 At2g3... 35 0.092
TC221656 similar to UP|Q9SVS9 (Q9SVS9) Peptide transporter-like ... 30 2.3
TC230140 similar to UP|O03296 (O03296) Cytochrome b (Fragment), ... 30 2.9
TC205661 29 3.8
BE022522 29 3.8
BG725765 homologue to SP|P08640|AMYH Glucoamylase S1/S2 precurso... 29 3.8
TC212052 29 3.8
TC210261 weakly similar to GB|CAA58476.1|929862|SCXORFS J0916 {S... 29 5.0
BM886161 28 8.6
TC232951 similar to UP|Q945B8 (Q945B8) Growth-on protein GRO10, ... 28 8.6
>TC219112 weakly similar to UP|Q94AA1 (Q94AA1) AT3g09470/F11F8_4, partial
(43%)
Length = 1150
Score = 437 bits (1125), Expect = e-123
Identities = 227/294 (77%), Positives = 248/294 (84%), Gaps = 1/294 (0%)
Frame = +3
Query: 168 NFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSDGQ-EGSTNGTTLLFVVFLSVMTFG 226
+ HEGAVIGDFNGEFW VY LHQFIGNLITFALLSD Q EGST GTTLLF+VFL +MTFG
Sbjct: 6 SLHEGAVIGDFNGEFWAVYALHQFIGNLITFALLSDNQQEGSTKGTTLLFIVFLFIMTFG 185
Query: 227 AILTCFLHKRGDYSKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQ 286
AIL CFL KR SKG + A G SLKSL +SL ALSDVKMLLIIPLIAYSGLQ
Sbjct: 186 AILMCFLRKRSANSKGQQELSGADAGACASLKSLSKSLASALSDVKMLLIIPLIAYSGLQ 365
Query: 287 HAFVWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFV 346
AFVWAEFTKYVVTP IG+SGVG +MA Y AFDGICSL+AGRLT GLTSIT+IVS G F
Sbjct: 366 QAFVWAEFTKYVVTPAIGISGVGSSMAAYAAFDGICSLLAGRLTSGLTSITTIVSAGLFA 545
Query: 347 QAVVLILLLLDFSMSSGFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQ 406
QAVVL+LLLL+FS+SSG +GT+YILFLA LLGIGDGVLMTQLNAL+G+LFKHD EGAFAQ
Sbjct: 546 QAVVLVLLLLNFSISSGLLGTVYILFLAGLLGIGDGVLMTQLNALIGILFKHDTEGAFAQ 725
Query: 407 LKIWQSATIAMVFFLAPYISFQAVIMVMLTLLCLSFCSFLWLALKVGNASSPST 460
LKIWQ ATIA+VFF AP ISF+AV+++ML LLC SFC FL LALKVG A SPST
Sbjct: 726 LKIWQCATIAIVFFFAPLISFKAVLVIMLALLCFSFCIFLLLALKVGKAPSPST 887
>BI787704 similar to GP|15146218|gb| AT3g09470/F11F8_4 {Arabidopsis
thaliana}, partial (14%)
Length = 421
Score = 67.0 bits (162), Expect(2) = 5e-20
Identities = 37/53 (69%), Positives = 44/53 (82%)
Frame = +3
Query: 9 DEETPLVVATDESPIQFQQHKSHTRDVHILSIAFLLIFLAYGAAQNLQSTLNT 61
DEETPLV D S ++ ++ HTRDVHILS+AFLL+FLA+GAAQNLQSTLNT
Sbjct: 144 DEETPLV--GDISGLEGRK-SHHTRDVHILSLAFLLVFLAFGAAQNLQSTLNT 293
Score = 48.9 bits (115), Expect(2) = 5e-20
Identities = 21/40 (52%), Positives = 26/40 (64%)
Frame = +2
Query: 75 GGFGYDFAWDIIFVFYIFFCVCFFGGADSRIQECFDCWNF 114
G GY WD +FVF+I CFFGGA SR+QE F W++
Sbjct: 296 G*LGYHLTWDPVFVFHIPLRGCFFGGASSRLQERFAYWDY 415
>TC219111
Length = 409
Score = 60.8 bits (146), Expect = 1e-09
Identities = 28/41 (68%), Positives = 33/41 (80%)
Frame = +2
Query: 420 FLAPYISFQAVIMVMLTLLCLSFCSFLWLALKVGNASSPST 460
F AP +SF+AV+++ML LLC SFC FL LALKVG SSPST
Sbjct: 2 FFAPLVSFKAVLVIMLALLCFSFCIFLLLALKVGKTSSPST 124
>TC231429 weakly similar to GB|AAP68235.1|31711758|BT008796 At2g36810
{Arabidopsis thaliana;} , partial (12%)
Length = 677
Score = 34.7 bits (78), Expect = 0.092
Identities = 28/113 (24%), Positives = 50/113 (43%), Gaps = 2/113 (1%)
Frame = +1
Query: 79 YDFAWDIIFVFYIFFCVCFFGGADSR-IQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCL 137
+D W II I+FC +D++ I + FG+LV + +V S L
Sbjct: 58 FDAPWPIIQANAIYFCSSMLSLSDNQHILAVYHSQVFGMLVGKLSRSPDAVVRATSSAAL 237
Query: 138 SWVLRF-YIMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLH 189
+L+ ++ W A + L ST+R+H +++ + +FW +Y H
Sbjct: 238 GLLLKSSHLCSWRAVELDRLESTSRNHDVESTKN--------*QQFWNIYQNH 372
>TC221656 similar to UP|Q9SVS9 (Q9SVS9) Peptide transporter-like protein,
partial (31%)
Length = 860
Score = 30.0 bits (66), Expect = 2.3
Identities = 26/112 (23%), Positives = 44/112 (39%), Gaps = 1/112 (0%)
Frame = +3
Query: 348 AVVLILLLLDFSMSSGF-IGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQ 406
A +LIL+ + + F +G +LFL ++G + ++ G ++ + GAF
Sbjct: 210 AAILILVNQGLATLAFFGVGVNLVLFLTRVMGQDNAEAANSVSKWTGTVYHFSLLGAFLS 389
Query: 407 LKIWQSATIAMVFFLAPYISFQAVIMVMLTLLCLSFCSFLWLALKVGNASSP 458
W + FQ + ++ L L LS FL GN P
Sbjct: 390 DSYWGRYMTCAI--------FQVIFVIGLVSLSLSSYIFLLKPSGCGNKELP 521
>TC230140 similar to UP|O03296 (O03296) Cytochrome b (Fragment), partial (5%)
Length = 1462
Score = 29.6 bits (65), Expect = 2.9
Identities = 19/71 (26%), Positives = 32/71 (44%), Gaps = 8/71 (11%)
Frame = +2
Query: 109 FDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVL--------RFYIMGWAACQGTYLTSTA 160
F+ F +++P F L + G+ ++W L ++GW A ++ S
Sbjct: 944 FESLIFTSIIVPQASFLGHLSGIVVGYAIAWGLIHGMSNYWALSLLGWIAL--VFVLSLK 1117
Query: 161 RSHAIDNNFHE 171
RS A+D NF E
Sbjct: 1118 RSGAVDLNFLE 1150
>TC205661
Length = 1383
Score = 29.3 bits (64), Expect = 3.8
Identities = 27/88 (30%), Positives = 45/88 (50%), Gaps = 1/88 (1%)
Frame = -3
Query: 349 VVLILLLLDFSMSSGFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLK 408
++L+LLLL F S+ F +++GI DGVLMT + A + + K ++ +
Sbjct: 766 LLLLLLLLPF*TST---------FTVSVMGITDGVLMTTVCAEVLVAVKFELSTNPSSSG 614
Query: 409 IWQSAT-IAMVFFLAPYISFQAVIMVML 435
W + T A+ FL+P +A +V L
Sbjct: 613 NWLTFTSAALGKFLSPSEDVEARELVNL 530
>BE022522
Length = 414
Score = 29.3 bits (64), Expect = 3.8
Identities = 26/89 (29%), Positives = 43/89 (48%), Gaps = 5/89 (5%)
Frame = -3
Query: 186 YTLHQFI---GNLITFALLSDGQEGSTNGTTLLFVVFLSVMTFGAILTCFLHK-RGDYSK 241
+ LHQ I GN ++ L+ + G N + + F FL + A + C L + RG+
Sbjct: 334 FDLHQIIRILGNKVSCMLVPNSARGE-N*SRVFFFFFLQKASL-AFVHCLLEEARGELVG 161
Query: 242 GGYKHLDAGTGQSKS-LKSLCRSLTGALS 269
G Y H+ G G + + ++L R G L+
Sbjct: 160 GEYDHVHDGEGHAAAHRRALRRRAAGNLA 74
>BG725765 homologue to SP|P08640|AMYH Glucoamylase S1/S2 precursor (EC
3.2.1.3) (Glucan 1 4-alpha- glucosidase), partial (1%)
Length = 389
Score = 29.3 bits (64), Expect = 3.8
Identities = 18/63 (28%), Positives = 28/63 (43%), Gaps = 2/63 (3%)
Frame = -2
Query: 81 FAWDIIFVFYIFFCVCFFGGADSRIQECFDCWNF--GLLVIPGCKFEAQLVYVGSGFCLS 138
F + I+ +F + F G A CW+ G++++P +V GSG C
Sbjct: 271 FVFAIVLLFALLFVTAVRGAAGGYDAAGCGCWSLLRGVVLLP-------VVTAGSGGCWV 113
Query: 139 WVL 141
WVL
Sbjct: 112 WVL 104
>TC212052
Length = 627
Score = 29.3 bits (64), Expect = 3.8
Identities = 17/51 (33%), Positives = 24/51 (46%), Gaps = 1/51 (1%)
Frame = +2
Query: 96 CFFGGADSRI-QECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFYI 145
C G D+R Q WN ++ + G V +GS CLS LR+Y+
Sbjct: 8 CSTGLLDARTAQSVITVWNGLIITVLGLGSVLGCVIIGSSSCLSSQLRYYV 160
>TC210261 weakly similar to GB|CAA58476.1|929862|SCXORFS J0916 {Saccharomyces
cerevisiae;} , partial (7%)
Length = 816
Score = 28.9 bits (63), Expect = 5.0
Identities = 25/81 (30%), Positives = 33/81 (39%)
Frame = +1
Query: 128 LVYVGSGFCLSWVLRFYIMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYT 187
LV VG FCL L Y++ S R I + EG + F +WG+Y
Sbjct: 55 LVLVGYQFCLVQGLNHYLL-----------SNERGMDIISQNKEG-IFSIFG--YWGMYL 192
Query: 188 LHQFIGNLITFALLSDGQEGS 208
L +GN + F S G S
Sbjct: 193 LGVHLGNYLIFGSHSSGFRSS 255
>BM886161
Length = 470
Score = 28.1 bits (61), Expect = 8.6
Identities = 11/24 (45%), Positives = 18/24 (74%)
Frame = -3
Query: 437 LLCLSFCSFLWLALKVGNASSPST 460
+ CLS CS L+LA+K+ ++S S+
Sbjct: 210 IFCLSICSILFLAIKLASSSYKSS 139
>TC232951 similar to UP|Q945B8 (Q945B8) Growth-on protein GRO10, partial
(29%)
Length = 502
Score = 28.1 bits (61), Expect = 8.6
Identities = 29/113 (25%), Positives = 46/113 (40%)
Frame = +1
Query: 335 SITSIVSFGAFVQAVVLILLLLDFSMSSGFIGTLYILFLAALLGIGDGVLMTQLNALLGM 394
++ + V F F + +L L MSS FI L +LF G L T L ALL
Sbjct: 142 NVKAAVLFVVFASCFLFMLYKL---MSSWFIDVLVVLFCIG----GIEGLQTCLVALLSR 300
Query: 395 LFKHDMEGAFAQLKIWQSATIAMVFFLAPYISFQAVIMVMLTLLCLSFCSFLW 447
FKH E + P++ + + + ++ C++F + LW
Sbjct: 301 WFKHAGES----------------YIKVPFLGAISYLTLAVSPFCITF-AILW 408
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.329 0.143 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,765,376
Number of Sequences: 63676
Number of extensions: 360884
Number of successful extensions: 3033
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 2999
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3028
length of query: 460
length of database: 12,639,632
effective HSP length: 101
effective length of query: 359
effective length of database: 6,208,356
effective search space: 2228799804
effective search space used: 2228799804
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC135797.3


