

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135795.6 - phase: 0
(267 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
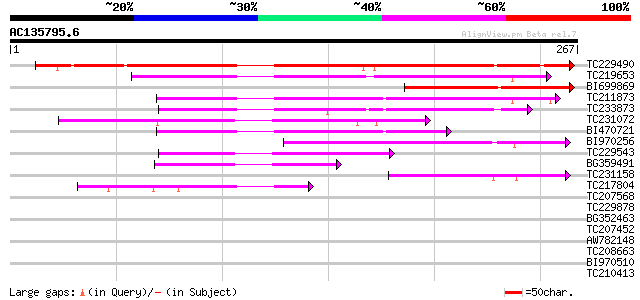
Score E
Sequences producing significant alignments: (bits) Value
TC229490 similar to UP|Q8S9M5 (Q8S9M5) At2g41130/T3K9.10, partia... 310 6e-85
TC219653 similar to GB|AAL79583.1|18958020|AY079033 AT3g25710/K1... 152 2e-37
BI699869 116 1e-26
TC211873 weakly similar to UP|Q6ZJC8 (Q6ZJC8) BHLH protein famil... 100 7e-22
TC233873 weakly similar to UP|Q6ZJC8 (Q6ZJC8) BHLH protein famil... 100 7e-22
TC231072 weakly similar to UP|Q9LGL7 (Q9LGL7) ESTs C26093(C11622... 88 3e-18
BI470721 82 3e-16
BI970256 73 1e-13
TC229543 similar to UP|Q9LGL7 (Q9LGL7) ESTs C26093(C11622), part... 71 6e-13
BG359491 57 9e-09
TC231158 51 5e-07
TC217804 41 5e-04
TC207568 similar to UP|Q9LSL1 (Q9LSL1) Arabidopsis thaliana geno... 38 0.005
TC229878 37 0.009
BG352463 similar to GP|11245494|gb| SPATULA {Arabidopsis thalian... 36 0.021
TC207452 similar to UP|Q9FUA4 (Q9FUA4) SPATULA, partial (31%) 35 0.035
AW782148 homologue to GP|11245494|gb| SPATULA {Arabidopsis thali... 35 0.046
TC208663 similar to UP|PIF4_ARATH (Q8W2F3) Phytochrome-interacti... 34 0.060
BI970510 34 0.060
TC210413 weakly similar to UP|Q6YZC9 (Q6YZC9) Uridylyl transfera... 33 0.13
>TC229490 similar to UP|Q8S9M5 (Q8S9M5) At2g41130/T3K9.10, partial (56%)
Length = 1006
Score = 310 bits (793), Expect = 6e-85
Identities = 177/265 (66%), Positives = 198/265 (73%), Gaps = 11/265 (4%)
Frame = +1
Query: 13 NNSELFQFL-------VANNPSFFEYSTPSSTMMQQSFCSSSDNNNNYYHPFEVSEITDT 65
+NS L QFL N FF ++ P + M ++S NN NYY PF +S+IT+T
Sbjct: 133 DNSHLLQFLPNTTTTTTTTNTPFF-HAAPHTHTMHHPLSTNSSNNTNYY-PFHLSQITET 306
Query: 66 PSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKS 125
PS DRALAA+KNHKEAEKRRRERINSHLDHLRTLLPCNSKT
Sbjct: 307 PSHHDRALAAMKNHKEAEKRRRERINSHLDHLRTLLPCNSKT-----------------D 435
Query: 126 KGSRLAKVVERVKELRQQTCQITQVETVPCEADGXTVISA-GSDMS---GEGRLIFKASL 181
K S LAKVV+RVKEL+QQT +IT++ETVP E D TV+S G D + G+GRLIFKASL
Sbjct: 436 KASLLAKVVQRVKELKQQTSEITELETVPSETDEITVLSTTGGDYASGGGDGRLIFKASL 615
Query: 182 CCEDRSDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAAEKEHNSIESIHFLQNSL 241
CCEDRSDLIPDLIEIL SLHLKTLKAEMATLGGRTRNVLVVAA+KEH SIESIHFLQNSL
Sbjct: 616 CCEDRSDLIPDLIEILNSLHLKTLKAEMATLGGRTRNVLVVAADKEH-SIESIHFLQNSL 792
Query: 242 RSLLDRSSGCNDRSKRRRGLDRRMM 266
RS+LDRSS DRSKRRRGLDRR+M
Sbjct: 793 RSILDRSSS-GDRSKRRRGLDRRLM 864
>TC219653 similar to GB|AAL79583.1|18958020|AY079033 AT3g25710/K13N2_1
{Arabidopsis thaliana;} , partial (47%)
Length = 1346
Score = 152 bits (384), Expect = 2e-37
Identities = 87/208 (41%), Positives = 126/208 (59%), Gaps = 10/208 (4%)
Frame = +2
Query: 58 EVSEITDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSS 117
E++++T + +ALAA K+H EAE+RRRERIN+HL LR+LLP +KT
Sbjct: 476 ELAKMTAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKT---------- 625
Query: 118 RTCERFKSKGSRLAKVVERVKELRQQTCQITQVETVPCEADGXTVISAGSDMSGEGRLIF 177
K S LA+V++ VKEL++QT I + VP EAD TV+ + +G +
Sbjct: 626 -------DKASLLAEVIQHVKELKRQTSLIAETSPVPTEADELTVVDEADE---DGNSVI 775
Query: 178 KASLCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAAEKEHNSIESIH-- 235
KASLCCEDRSDL P+LI+ LK+L L+TLKAE+ TLGGR +NVL + E+ +S H
Sbjct: 776 KASLCCEDRSDLFPELIKTLKALRLRTLKAEITTLGGRVKNVLFITGEETDSSSSEDHSQ 955
Query: 236 --------FLQNSLRSLLDRSSGCNDRS 255
+Q +L++++++S G + S
Sbjct: 956 QQQQCCISSIQEALKAVMEKSVGDHHES 1039
>BI699869
Length = 421
Score = 116 bits (290), Expect = 1e-26
Identities = 61/80 (76%), Positives = 71/80 (88%)
Frame = +1
Query: 187 SDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAAEKEHNSIESIHFLQNSLRSLLD 246
SDLIP+LIEIL+SL LKTLKAEMATLGGRTRNVLVVA +K+H+ ESI FLQNSL+SL++
Sbjct: 1 SDLIPELIEILRSLRLKTLKAEMATLGGRTRNVLVVATDKDHSG-ESIQFLQNSLKSLVE 177
Query: 247 RSSGCNDRSKRRRGLDRRMM 266
RSS NDRSKRRR LDR+ +
Sbjct: 178 RSSNSNDRSKRRRVLDRKFI 237
>TC211873 weakly similar to UP|Q6ZJC8 (Q6ZJC8) BHLH protein family-like,
partial (25%)
Length = 1149
Score = 100 bits (249), Expect = 7e-22
Identities = 67/199 (33%), Positives = 108/199 (53%), Gaps = 9/199 (4%)
Frame = +3
Query: 70 DRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSR 129
+R++ ALK+H EAE++RR RIN+HLD LR+++P K K S
Sbjct: 357 ERSIEALKSHSEAERKRRARINAHLDTLRSVIPGAMKM-----------------DKASL 485
Query: 130 LAKVVERVKELRQQTCQITQVETVPCEADGXTVISAGSDMSGEGRLIFKASLCCEDRSDL 189
L +V+ +KEL++ Q + +P + D +V ++G I +ASLCCE + L
Sbjct: 486 LGEVIRHLKELKKNAAQACEGLMIPKDNDEISVEEQEGGLNGFPYSI-RASLCCEYKPGL 662
Query: 190 IPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAAEKEHNSIESIH------FLQNSLRS 243
+ D+ + L +LHL +A++ATL GR +NV V+ + KE N ++ + + +L+S
Sbjct: 663 LSDIKQALDALHLMITRADIATLEGRMKNVFVIISCKEQNFEDAAYRQFLAVSVHQALKS 842
Query: 244 LLDRSSGCND---RSKRRR 259
+L+R S D KRRR
Sbjct: 843 VLNRFSVSEDILGTRKRRR 899
>TC233873 weakly similar to UP|Q6ZJC8 (Q6ZJC8) BHLH protein family-like,
partial (25%)
Length = 817
Score = 100 bits (249), Expect = 7e-22
Identities = 68/193 (35%), Positives = 105/193 (54%), Gaps = 17/193 (8%)
Frame = +3
Query: 71 RALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRL 130
++ A K+H+EAE+RRR+RINSHL LRTLLP +K+ K S L
Sbjct: 123 KSTEACKSHREAERRRRQRINSHLSTLRTLLPNAAKS-----------------DKASLL 251
Query: 131 AKVVERVKELRQQTCQIT-----------------QVETVPCEADGXTVISAGSDMSGEG 173
+VVE VK LR+Q +T + P E D TV + GE
Sbjct: 252 GEVVEHVKRLRKQADDVTCGDSYSSRSGEPGSVRSEAWPFPGECDEVTVSYCDGE-DGEP 428
Query: 174 RLIFKASLCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAAEKEHNSIES 233
+ + KA++CC DR+ L D+ + ++S+ K ++AEM T+GGRT++V+VV EKE E
Sbjct: 429 KRV-KATVCCGDRTGLNRDVSQAIRSVRAKAVRAEMMTVGGRTKSVVVVEWEKEE---EE 596
Query: 234 IHFLQNSLRSLLD 246
+ L+ +L+++++
Sbjct: 597 VGALERALKAVVE 635
>TC231072 weakly similar to UP|Q9LGL7 (Q9LGL7) ESTs C26093(C11622), partial
(21%)
Length = 590
Score = 88.2 bits (217), Expect = 3e-18
Identities = 57/186 (30%), Positives = 93/186 (49%), Gaps = 11/186 (5%)
Frame = +1
Query: 24 NNPSFFEYSTPSSTMMQQSFCSSSDNNNNYYHPFEVSEITDTPSQ---QDRALAALKNHK 80
N+ +S P++T ++S ++ + E P+ +DRA +A K+H
Sbjct: 79 NSDQLSPFSVPTTTQQTLPLVATSTSDPAAAASLQFGEFPSWPAPIAAEDRAASASKSHS 258
Query: 81 EAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERVKEL 140
+AEKRRR+RIN+ L LR L+P + K K + L VV+ VK+L
Sbjct: 259 QAEKRRRDRINAQLATLRKLIPMSDK-----------------MDKATLLGSVVDHVKDL 387
Query: 141 RQQTCQITQVETVPCEADGXTV-ISAGSDMSG-------EGRLIFKASLCCEDRSDLIPD 192
+++ +++ TVP E D T+ D S + +I KAS+CC+DR +L P+
Sbjct: 388 KRKAMDVSKAITVPTETDEVTIDYHQAQDESYTKKVNILKENIIIKASVCCDDRPELFPE 567
Query: 193 LIEILK 198
LI++LK
Sbjct: 568 LIQVLK 585
>BI470721
Length = 421
Score = 81.6 bits (200), Expect = 3e-16
Identities = 49/139 (35%), Positives = 76/139 (54%)
Frame = +1
Query: 70 DRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSR 129
+R++ ALKNH EAE+RRR RIN+HLD LR+++P K K +
Sbjct: 58 ERSIEALKNHSEAERRRRARINAHLDTLRSVIPGAKKL-----------------DKATL 186
Query: 130 LAKVVERVKELRQQTCQITQVETVPCEADGXTVISAGSDMSGEGRLIFKASLCCEDRSDL 189
L +V+ +KEL+ Q ++ +P ++D V ++G I KASLCCE + L
Sbjct: 187 LGEVIRHLKELKTNATQASEGLMIPKDSDEIRVEEQEGGLNGFPYSI-KASLCCEYKPGL 363
Query: 190 IPDLIEILKSLHLKTLKAE 208
+ D+ + L +LHL ++AE
Sbjct: 364 LTDIRQALDALHLMIIRAE 420
>BI970256
Length = 703
Score = 73.2 bits (178), Expect = 1e-13
Identities = 46/144 (31%), Positives = 79/144 (53%), Gaps = 9/144 (6%)
Frame = -3
Query: 130 LAKVVERVKELRQQTCQITQVETVPCEADGXTVISAGSDMSGEGRLIFKASLCCEDRSDL 189
LA+V+ +VKEL++ +++ +P +AD V GEG + + A++CC+ R ++
Sbjct: 674 LAEVISQVKELKKNXXXVSKGFLIPKDADEVKVEPYNDHEGGEGSMSYSATICCDFRPEI 495
Query: 190 IPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAAEKEHNSIESIHF---------LQNS 240
+ DL + L SL L +KAE++TL GR +NV V KE +I +I F + +
Sbjct: 494 LXDLRQTLDSLPLHLVKAEISTLAGRMKNVFVFPCCKE--NINNIDFEKCQALASSVPQA 321
Query: 241 LRSLLDRSSGCNDRSKRRRGLDRR 264
L S+++++S D S R +R
Sbjct: 320 LCSVMEKASASLDFSPRTSHASKR 249
>TC229543 similar to UP|Q9LGL7 (Q9LGL7) ESTs C26093(C11622), partial (24%)
Length = 1149
Score = 70.9 bits (172), Expect = 6e-13
Identities = 41/111 (36%), Positives = 60/111 (53%)
Frame = +2
Query: 71 RALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRL 130
+ALAALKNH EAE+RRRERIN HL LR L+P K K + L
Sbjct: 866 KALAALKNHSEAERRRRERINGHLATLRGLVPSTEK-----------------MDKATLL 994
Query: 131 AKVVERVKELRQQTCQITQVETVPCEADGXTVISAGSDMSGEGRLIFKASL 181
A+V+ +VKEL++ ++++ +P +AD V GEG + + A++
Sbjct: 995 AEVISQVKELKKNAAEVSKGFLIPKDADEVKVEPYNDHEGGEGSMSYSATI 1147
>BG359491
Length = 361
Score = 57.0 bits (136), Expect = 9e-09
Identities = 30/88 (34%), Positives = 52/88 (59%)
Frame = +2
Query: 69 QDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGS 128
++RA++A K+H++AEKRRR+RIN+ L LR L+P + K K +
Sbjct: 146 EERAISASKSHRQAEKRRRDRINAQLATLRKLIPKSDK-----------------MDKAA 274
Query: 129 RLAKVVERVKELRQQTCQITQVETVPCE 156
L V+++VK+L+++ +++ TVP E
Sbjct: 275 LLGSVIDQVKDLKRKAMDVSRAFTVPTE 358
>TC231158
Length = 642
Score = 51.2 bits (121), Expect = 5e-07
Identities = 34/93 (36%), Positives = 51/93 (54%), Gaps = 7/93 (7%)
Frame = +1
Query: 179 ASLCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAAEK---EHNSIESIH 235
A++CC+ RS+++ DL + L SL L +KAE++TL GR +NV V K + IE
Sbjct: 16 ATICCDFRSEILSDLRQALDSLPLHLVKAEISTLAGRMKNVFVFTCCKGNINNIDIEKCQ 195
Query: 236 FL----QNSLRSLLDRSSGCNDRSKRRRGLDRR 264
L +L S+LD++S D S R +R
Sbjct: 196 ALASTVHQALCSVLDKASATLDFSPRTSHASKR 294
>TC217804
Length = 1578
Score = 41.2 bits (95), Expect = 5e-04
Identities = 31/124 (25%), Positives = 59/124 (47%), Gaps = 13/124 (10%)
Frame = +1
Query: 33 TPSSTMMQQSFCS---SSDNNNNYYHPFEVSEITDTP---SQQDRALAALKN-------H 79
+P +T C+ ++D+ ++ H E+ D ++ DR+ + K H
Sbjct: 646 SPENTTCSGKQCTGTTTNDDRDSISHRISQGEVPDEDYKATKVDRSSGSNKRIKANSVVH 825
Query: 80 KEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERVKE 139
K++E+RRR++IN + L+ L+P +SKT K S L +V++ +K+
Sbjct: 826 KQSERRRRDKINQRMKELQKLVPNSSKT-----------------DKASMLDEVIQYMKQ 954
Query: 140 LRQQ 143
L+ Q
Sbjct: 955 LQAQ 966
>TC207568 similar to UP|Q9LSL1 (Q9LSL1) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K21L13, partial (50%)
Length = 914
Score = 37.7 bits (86), Expect = 0.005
Identities = 38/174 (21%), Positives = 75/174 (42%), Gaps = 1/174 (0%)
Frame = +1
Query: 82 AEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERVKELR 141
AE+RRR+R+N L LR+++P SK + +S T + K R+ K+ E +E+
Sbjct: 142 AERRRRKRLNDRLSMLRSIVPKISKMDR---TSILGDTIDYMKELLERIGKLQE--EEIE 306
Query: 142 QQTCQITQVETVPCEADGXTVISAGSDMSGEGR-LIFKASLCCEDRSDLIPDLIEILKSL 200
+ T QI + ++ E R + S+CC + L+ + L++L
Sbjct: 307 EGTNQINLLGISKELKPNEVMVRNSPKFDVERRDQDTRISICCATKPGLLLSTVNTLEAL 486
Query: 201 HLKTLKAEMATLGGRTRNVLVVAAEKEHNSIESIHFLQNSLRSLLDRSSGCNDR 254
L+ + +++ + ++ N + Q ++ L R++G R
Sbjct: 487 GLEIHQCVISSFNDFSMQASCTEVAEQRNCMS-----QEEIKQALFRNAGYGGR 633
>TC229878
Length = 509
Score = 37.0 bits (84), Expect = 0.009
Identities = 24/80 (30%), Positives = 35/80 (43%)
Frame = +2
Query: 64 DTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERF 123
D PS R H E E+RRR +IN LR L+P N + R
Sbjct: 110 DEPSTGKRVNPQRSKHSETEQRRRSKINERFQVLRDLIPQNDQKR--------------- 244
Query: 124 KSKGSRLAKVVERVKELRQQ 143
K S L +V+E ++ L+++
Sbjct: 245 -DKASFLLEVIEYIQFLQEK 301
>BG352463 similar to GP|11245494|gb| SPATULA {Arabidopsis thaliana}, partial
(19%)
Length = 471
Score = 35.8 bits (81), Expect = 0.021
Identities = 23/78 (29%), Positives = 36/78 (45%)
Frame = +3
Query: 66 PSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKS 125
P + A + H +EKRRR RIN + L+ L+P ++KT
Sbjct: 24 PRSSSKRSRAAEFHNLSEKRRRSRINEKMKALQNLIPNSNKT-----------------D 152
Query: 126 KGSRLAKVVERVKELRQQ 143
K S L + +E +K+L+ Q
Sbjct: 153 KASMLDEAIEYLKQLQLQ 206
>TC207452 similar to UP|Q9FUA4 (Q9FUA4) SPATULA, partial (31%)
Length = 1207
Score = 35.0 bits (79), Expect = 0.035
Identities = 31/120 (25%), Positives = 52/120 (42%), Gaps = 11/120 (9%)
Frame = +2
Query: 35 SSTMMQQSFCSSSDNNNNYYHPFEVSEITDTPSQQ-----------DRALAALKNHKEAE 83
S+ + S S+N N+ Y E E + P+++ + A + H +E
Sbjct: 2 SAANVSSSSAGVSENENDDYD-CESEEGDEAPAEEVPTKAASARSSSKRSRAAEVHN*SE 178
Query: 84 KRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERVKELRQQ 143
KRRR RIN + L+ L+P ++KT K S L + +E +K+L+ Q
Sbjct: 179 KRRRGRINEKMKALQNLIPNSNKT-----------------DKASMLDEAIEYLKQLQLQ 307
>AW782148 homologue to GP|11245494|gb| SPATULA {Arabidopsis thaliana},
partial (17%)
Length = 381
Score = 34.7 bits (78), Expect = 0.046
Identities = 32/125 (25%), Positives = 54/125 (42%), Gaps = 10/125 (8%)
Frame = +1
Query: 29 FEYSTPSSTMMQQSFCSSSDNNNNYYHPFE------VSEITDTPSQQDRALA----ALKN 78
F+ S+ + S S+N N+ Y ++E T + R+ + A +
Sbjct: 49 FKGQGASAANVSSSSAGVSENENDDYDCESEEGVEALAEEVPTKAASSRSSSKRSRAAEV 228
Query: 79 HKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERVK 138
H +EKRRR RIN + L+ L+P ++KT K S L + +E +K
Sbjct: 229 HNLSEKRRRSRINEKMKALQNLIPNSNKT-----------------DKASMLDEAIEYLK 357
Query: 139 ELRQQ 143
+L+ Q
Sbjct: 358 QLQLQ 372
>TC208663 similar to UP|PIF4_ARATH (Q8W2F3) Phytochrome-interacting factor 4
(Basic helix-loop-helix protein 9) (bHLH9) (AtbHLH009)
(Short under red-light 2), partial (19%)
Length = 1102
Score = 34.3 bits (77), Expect = 0.060
Identities = 22/69 (31%), Positives = 34/69 (48%)
Frame = +1
Query: 75 ALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVV 134
A + H +E+RRR+RIN + L+ L+P +SKT K S L + +
Sbjct: 262 AAEVHNLSERRRRDRINEKMKALQQLIPHSSKT-----------------DKASMLEEAI 390
Query: 135 ERVKELRQQ 143
E +K L+ Q
Sbjct: 391 EYLKSLQLQ 417
>BI970510
Length = 646
Score = 34.3 bits (77), Expect = 0.060
Identities = 26/70 (37%), Positives = 37/70 (52%), Gaps = 9/70 (12%)
Frame = -1
Query: 199 SLHLKTLKAEMATLGGRTRNVLVVAAEKEHNSIESIHFLQ-------NSLRSLLDRSSGC 251
+L L + E+ATLGG +V V+ + KE N IE + Q + RS+LDR S
Sbjct: 637 ALXLMIIXXEIATLGGXXNSVFVIISCKEQN-IEDPEYRQXXAGSVHQAXRSVLDRFSVS 461
Query: 252 ND--RSKRRR 259
D S++RR
Sbjct: 460 QDMLESRKRR 431
>TC210413 weakly similar to UP|Q6YZC9 (Q6YZC9) Uridylyl transferase-like,
partial (25%)
Length = 695
Score = 33.1 bits (74), Expect = 0.13
Identities = 17/45 (37%), Positives = 23/45 (50%)
Frame = +3
Query: 178 KASLCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVV 222
K LC EDR L+ D+ I + L +AE+ T G + NV V
Sbjct: 96 KLELCGEDRVGLLSDVTRIFRENGLSVNRAEVTTRGSQAMNVFYV 230
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,201,873
Number of Sequences: 63676
Number of extensions: 140722
Number of successful extensions: 1141
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 1121
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1126
length of query: 267
length of database: 12,639,632
effective HSP length: 96
effective length of query: 171
effective length of database: 6,526,736
effective search space: 1116071856
effective search space used: 1116071856
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC135795.6


