

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135795.1 - phase: 0
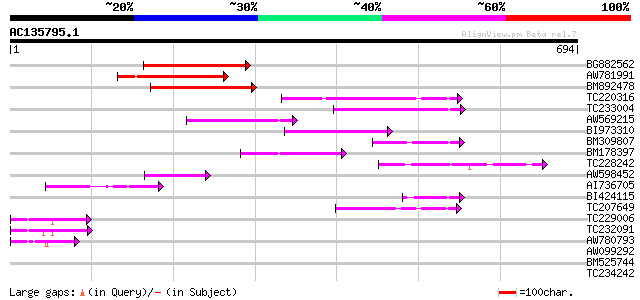
(694 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG882562 170 2e-42
AW781991 140 2e-33
BM892478 138 7e-33
TC220316 119 3e-27
TC233004 weakly similar to PIR|G96565|G96565 F6D8.26 [imported] ... 105 5e-23
AW569215 weakly similar to GP|4914326|gb| F14N23.12 {Arabidopsis... 104 1e-22
BI973310 99 6e-21
BM309807 similar to GP|5764395|gb| far-red impaired response pro... 86 4e-17
BM178397 similar to GP|4914326|gb| F14N23.12 {Arabidopsis thalia... 80 4e-15
TC228242 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired r... 75 1e-13
AW598452 weakly similar to GP|15982769|gb| At2g27110/T20P8.16 {A... 58 2e-08
AI736705 similar to GP|11994736|db far-red impaired response pro... 54 2e-07
BI424115 similar to GP|15982769|gb| At2g27110/T20P8.16 {Arabidop... 53 5e-07
TC207649 similar to UP|Q9SY66 (Q9SY66) F14N23.12 (At1g10240/F14N... 47 2e-05
TC229006 47 3e-05
TC232091 45 1e-04
AW780793 42 7e-04
AW099292 39 0.008
BM525744 39 0.010
TC234242 33 0.42
>BG882562
Length = 416
Score = 170 bits (430), Expect = 2e-42
Identities = 75/130 (57%), Positives = 98/130 (74%)
Frame = +3
Query: 165 GDAEAIQNYFVRMQKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYL 224
GD E I NYF R+Q N F+YVMD++D +LRNVFW+D+R RAAY YFG+V+ FD+T L
Sbjct: 27 GDPELISNYFCRIQLMNPNFFYVMDLNDDGQLRNVFWIDSRSRAAYSYFGDVVAFDSTCL 206
Query: 225 TNKYDMPFAPFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQD 284
+N Y++P FVGVNHHG+SVLLGC LL++E +T+ WLF+ WL CM GR P IIT+
Sbjct: 207 SNNYEIPLVAFVGVNHHGKSVLLGCGLLADETFETYIWLFRAWLTCMTGRPPQTIITNXC 386
Query: 285 RAMKKAIEDV 294
+AM+ AI +V
Sbjct: 387 KAMQSAIAEV 416
>AW781991
Length = 419
Score = 140 bits (353), Expect = 2e-33
Identities = 63/136 (46%), Positives = 86/136 (62%)
Frame = +3
Query: 132 SMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEAIQNYFVRMQKKNSQFYYVMDVD 191
+++ E G + F E DCRNY+ + RLR GD + + +Y +M +N F+Y + D
Sbjct: 15 ALIKEYGGISKVGFTEVDCRNYM-RNNRLRSLEGDIQLVLDYLRQMHAENPNFFYAVQGD 191
Query: 192 DKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGVNHHGQSVLLGCAL 251
+ + NVFW D + R Y +FG+ +TFDTTY +N+Y +PFA F GVNHHGQ VL GCA
Sbjct: 192 EDQSITNVFWADPKARMNYTFFGDTVTFDTTYRSNRYRLPFAFFTGVNHHGQPVLFGCAF 371
Query: 252 LSNEDTKTFSWLFKTW 267
L NE +F WLFKTW
Sbjct: 372 LINESEASFVWLFKTW 419
>BM892478
Length = 429
Score = 138 bits (348), Expect = 7e-33
Identities = 63/130 (48%), Positives = 90/130 (68%)
Frame = +3
Query: 173 YFVRMQKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPF 232
YF Q +++ F+Y M+VD+ + + N+FW D R R + +FG+V+ DT+Y Y +PF
Sbjct: 39 YFQSRQAEDTGFFYAMEVDNGNCM-NIFWADGRSRYSCSHFGDVLVLDTSYRKTVYLVPF 215
Query: 233 APFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAIE 292
A FVGVNHH Q VLLGCAL+++E ++F+WLF+TWL M GR P +I DQD A+++AI
Sbjct: 216 ATFVGVNHHKQPVLLGCALIADESEESFTWLFQTWLRAMSGRLPLTVIADQDIAIQRAIA 395
Query: 293 DVFPKARHRW 302
VFP HR+
Sbjct: 396 KVFPVTHHRF 425
>TC220316
Length = 988
Score = 119 bits (299), Expect = 3e-27
Identities = 69/222 (31%), Positives = 115/222 (51%)
Frame = +1
Query: 333 YDSSSISDFMEKWKKMIECYELHDNEWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESM 392
Y S ++ + W Y L D+ WLK ++ +R WVP+Y++ TF+AG+ +ES+
Sbjct: 10 YQSQTVDEXXATWNXXXNKYGLKDDAWLKEMYQKRASWVPLYLKGTFFAGI---PMNESL 180
Query: 393 NSFFDGYVSSKTTLKQFVEQYDNALKDKIEKESMADFVSFNTTIACISLFGFESQFQKAF 452
+SFF ++++T L +F+ +Y+ L+ + E+E DF + N + E Q ++ +
Sbjct: 181 DSFFGALLNAQTPLMEFIPRYERGLERRREEERKEDFNTSNFQPILQTKEPVEEQCRRLY 360
Query: 453 TNAKFKEFQIEIASMIYCNAFLEGMENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEI 512
T FK FQ E+ F E S + V ++ V FN + I
Sbjct: 361 TLTVFKIFQKELLQCFSYLGFKIFEEGGLSRYMVRRCGNDMEKHV-----VTFNASNISI 525
Query: 513 QCACCLFEFKGILCRHILCVLQLTGKTESVPSCYILSRWRKD 554
C+C +FE++G+LCRH+L V Q+ + VPS YIL RW ++
Sbjct: 526 SCSCQMFEYEGVLCRHVLRVFQIL-QLREVPSRYILHRWTRN 648
>TC233004 weakly similar to PIR|G96565|G96565 F6D8.26 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(8%)
Length = 551
Score = 105 bits (263), Expect = 5e-23
Identities = 62/163 (38%), Positives = 87/163 (53%), Gaps = 2/163 (1%)
Frame = +1
Query: 397 DGYVSSKTTLKQFVEQYDNALKDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAK 456
DGYV T+ K+FV++YD L K KE+MAD + N + + FE Q K +T
Sbjct: 1 DGYVHKHTSFKEFVDKYDLVLHRKHLKEAMADLETRNVSFELKTRCNFEVQLAKVYTKEI 180
Query: 457 FKEFQIEIASMIYC-NAFLEGMENLNSTFCVIESKKVYDRFKDTR-FRVIFNEKDFEIQC 514
F++FQ E+ M C N + T+ V E +V K + F V++ + +I+C
Sbjct: 181 FQKFQSEVEGMYSCFNTRQVSVNGSIITYVVKERVEVEGNEKGVKSFEVLYETTELDIRC 360
Query: 515 ACCLFEFKGILCRHILCVLQLTGKTESVPSCYILSRWRKDIKR 557
C LF +KG LCR L VL G E +PS YIL RWR+D K+
Sbjct: 361 ICSLFNYKGYLCRRALNVLNYNG-IEEIPSRYILHRWRRDFKQ 486
>AW569215 weakly similar to GP|4914326|gb| F14N23.12 {Arabidopsis thaliana},
partial (19%)
Length = 425
Score = 104 bits (260), Expect = 1e-22
Identities = 49/138 (35%), Positives = 81/138 (58%), Gaps = 2/138 (1%)
Frame = +1
Query: 217 ITFDTTYLTNKYDMPFAPFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAP 276
+ FDTTY YDM ++GV+++G + CALL +E+ ++FSW K +L M G+AP
Sbjct: 10 VVFDTTYRVEAYDMLLGIWLGVDNNGMTCFFSCALLRDENIQSFSWALKAFLGFMKGKAP 189
Query: 277 NAIITDQDRAMKKAIEDVFPKARHRWCLWHLMKKVPE--KLGRHSHYESIKLLLHDAVYD 334
I+TD + +K+AI P+ +H +C+WH++ K + L S Y+ K H +Y+
Sbjct: 190 QTILTDHNMWLKEAIAVELPQTKHAFCIWHILSKFSDWFSLLLGSQYDEWKAEFH-RLYN 366
Query: 335 SSSISDFMEKWKKMIECY 352
+ DF E W++M++ Y
Sbjct: 367 LEQVEDFEEGWRQMVDQY 420
>BI973310
Length = 457
Score = 99.0 bits (245), Expect = 6e-21
Identities = 48/132 (36%), Positives = 77/132 (57%)
Frame = +3
Query: 337 SISDFMEKWKKMIECYELHDNEWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFF 396
+I +F W ++E + + DNEWL+ +++ R WVPVY+R+TF+ +S + +E + SFF
Sbjct: 3 TIDEFESYWHSLLERFYVMDNEWLQSIYNSRQHWVPVYLRETFFGEISLNEGNEYLISFF 182
Query: 397 DGYVSSKTTLKQFVEQYDNALKDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAK 456
DGYV S TTL+ V QY+ A+ EKE AD+ + N++ + E Q +T
Sbjct: 183 DGYVFSSTTLQVLVRQYEKAVSSWHEKELKADYDTTNSSPVLKTPSPMEKQAASLYTRKI 362
Query: 457 FKEFQIEIASMI 468
F +FQ E+ +
Sbjct: 363 FMKFQEELVETL 398
>BM309807 similar to GP|5764395|gb| far-red impaired response protein
{Arabidopsis thaliana}, partial (15%)
Length = 428
Score = 86.3 bits (212), Expect = 4e-17
Identities = 48/112 (42%), Positives = 61/112 (53%)
Frame = +3
Query: 445 ESQFQKAFTNAKFKEFQIEIASMIYCNAFLEGMENLNSTFCVIESKKVYDRFKDTRFRVI 504
E Q +T+A FK+FQ+E+ + C + +E + + F V D KD F V
Sbjct: 48 EKQMSTVYTHAIFKKFQVEVLGVAGCQSRIEAGDGTIAKFIV------QDYEKDEEFLVT 209
Query: 505 FNEKDFEIQCACCLFEFKGILCRHILCVLQLTGKTESVPSCYILSRWRKDIK 556
+NE E+ C C LFE+KG LCRH L VLQ G VPS YIL RW KD K
Sbjct: 210 WNELSSEVSCFCRLFEYKGFLCRHALSVLQRCG-CSCVPSHYILKRWTKDAK 362
>BM178397 similar to GP|4914326|gb| F14N23.12 {Arabidopsis thaliana}, partial
(20%)
Length = 422
Score = 79.7 bits (195), Expect = 4e-15
Identities = 41/132 (31%), Positives = 70/132 (52%), Gaps = 2/132 (1%)
Frame = +1
Query: 283 QDRAMKKAIEDVFPKARHRWCLWHLMKKVPEKLGR--HSHYESIKLLLHDAVYDSSSISD 340
Q+ +K+A+ P +H +C+W ++ K P Y K + +Y+ S+ D
Sbjct: 1 QNICLKEALSTEMPTTKHAFCIWMIVAKFPSWFNAVLGERYNDWKAEFY-RLYNLESVED 177
Query: 341 FMEKWKKMIECYELHDNEWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYV 400
F W++M + LH N + L+ R W ++R F AGM+TT +S+S+N+F ++
Sbjct: 178 FELGWREMACSFGLHSNRHMVNLYSSRSLWALPFLRSHFLAGMTTTGQSKSINAFIQRFL 357
Query: 401 SSKTTLKQFVEQ 412
S++T L FVEQ
Sbjct: 358 SAQTRLAHFVEQ 393
>TC228242 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired response
protein; Mutator-like transposase-like protein;
phytochrome A signaling protein-like, partial (25%)
Length = 1354
Score = 75.1 bits (183), Expect = 1e-13
Identities = 57/228 (25%), Positives = 95/228 (41%), Gaps = 21/228 (9%)
Frame = +2
Query: 452 FTNAKFKEFQIEIASMIYCNAFLEGMENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFE 511
FT+A FK+ Q E+ + C+ + ++ +V+D + F V+ N+ E
Sbjct: 74 FTHAVFKKIQAEVIGAVACHPKADRQDDTTIVH------RVHDMETNKDFFVVVNQVKSE 235
Query: 512 IQCACCLFEFKGILCRHILCVLQLTGKTESVPSCYILSRWRKDIKRRHTL---------- 561
+ C C LFE++G LCRH L VLQ +G++ PS YIL RW KD K R+ +
Sbjct: 236 LSCICRLFEYRGYLCRHALFVLQYSGQS-VFPSQYILKRWTKDAKVRNIMGEESEHMLTR 412
Query: 562 -----------IKCGFDGNVELQLVGKACDAFYEVVSVGIHTEDDLLKVMNRINELKIEL 610
+K +G++ + G A A +E + V N E
Sbjct: 413 VQRYNDLCQRALKLSEEGSLSQESYGIAFHALHE-------AHKSCVSVNNSSKSSPTEA 571
Query: 611 NCEGSSSVIIEEDSLVQNQMAKILDPATTRSKGRPPSKRKASKLDQIV 658
G+ + E+ M+K + K P K+K + +++
Sbjct: 572 GTPGAHGQLSTEEDTQSRNMSK------SNKKKNPTKKKKVNSEAEVI 697
>AW598452 weakly similar to GP|15982769|gb| At2g27110/T20P8.16 {Arabidopsis
thaliana}, partial (7%)
Length = 256
Score = 57.8 bits (138), Expect = 2e-08
Identities = 25/80 (31%), Positives = 44/80 (54%)
Frame = +2
Query: 166 DAEAIQNYFVRMQKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLT 225
DA + Y RMQ +N +Y + +D+ R+ NV R R +Y++ G+ + D Y
Sbjct: 17 DAHNLLEYCKRMQAENHGCFYAIQLDEDDRVSNVL*AGTRSRTSYKHMGDAESLDAAYGK 196
Query: 226 NKYDMPFAPFVGVNHHGQSV 245
++Y + +AP+ G N HG+ +
Sbjct: 197 HRYAVSYAPYTGRNCHGRMI 256
>AI736705 similar to GP|11994736|db far-red impaired response protein;
Mutator-like transposase-like protein; phytochrome A
signaling, partial (14%)
Length = 383
Score = 54.3 bits (129), Expect = 2e-07
Identities = 38/144 (26%), Positives = 65/144 (44%)
Frame = +3
Query: 45 RARSYVSNSKNLLKPNPTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRC 104
RAR +S+N + C A +++ DGK I EHNH L P ++
Sbjct: 21 RARQNKQDSENSTGRRSCSKTDCKASMHVKRRSDGKWVIHSFVKEHNHELLPAQA----- 185
Query: 105 NKNLDPHTKRRLDINDQAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGT 164
+++Q R + +M + Y + G K+ +N DK R L L +
Sbjct: 186 -------------VSEQ----TRRMYAAMARQFAEYKTVV-GLKNEKNPFDKGRNLGLES 311
Query: 165 GDAEAIQNYFVRMQKKNSQFYYVM 188
G+A+ + ++F++MQ NS F+Y +
Sbjct: 312 GEAKLMLDFFIQMQNMNSNFFYAV 383
>BI424115 similar to GP|15982769|gb| At2g27110/T20P8.16 {Arabidopsis
thaliana}, partial (9%)
Length = 402
Score = 52.8 bits (125), Expect = 5e-07
Identities = 29/78 (37%), Positives = 44/78 (56%), Gaps = 2/78 (2%)
Frame = +2
Query: 481 NSTFCVIESKKVYDRFKDTR--FRVIFNEKDFEIQCACCLFEFKGILCRHILCVLQLTGK 538
NSTF V +F+D + + V N + + C+C +FE+ GILC+HIL V +T
Sbjct: 5 NSTFRVA-------KFEDDQKAYMVTLNHSELKANCSCQMFEYAGILCKHILTVFTVT-N 160
Query: 539 TESVPSCYILSRWRKDIK 556
++P YIL RW ++ K
Sbjct: 161 VLTLPPHYILKRWTRNAK 214
>TC207649 similar to UP|Q9SY66 (Q9SY66) F14N23.12 (At1g10240/F14N23_12),
partial (30%)
Length = 971
Score = 47.4 bits (111), Expect = 2e-05
Identities = 44/156 (28%), Positives = 67/156 (42%), Gaps = 1/156 (0%)
Frame = +2
Query: 399 YVSSKTTLKQFVEQYDNALKDKIEKESMADFVSFNTTIACISLFG-FESQFQKAFTNAKF 457
++S++T L FVEQ A+ K ++ + N C+ ES T F
Sbjct: 116 FLSAQTRLAHFVEQVAVAVDFK-DQTG*QQTMQQNLQNVCLKTGAPMESHAATILTPFAF 292
Query: 458 KEFQIEIASMIYCNAFLEGMENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACC 517
+ Q ++ + +F ++ F V K K V + ++ I C+C
Sbjct: 293 SKLQEQLVLAAHYASF-----SIEDGFLVRHHTKAEGGRK-----VYWAPQEGIISCSCH 442
Query: 518 LFEFKGILCRHILCVLQLTGKTESVPSCYILSRWRK 553
FEF GILCRH L VL TG +P Y+ RWR+
Sbjct: 443 QFEFTGILCRHSLRVLS-TGNCFQIPDRYLPIRWRR 547
>TC229006
Length = 991
Score = 47.0 bits (110), Expect = 3e-05
Identities = 35/108 (32%), Positives = 55/108 (50%), Gaps = 8/108 (7%)
Frame = +2
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINS--KNAKDGKKYFT-LGCTRARSYVS---NSK 54
M F SE+ +Y YAR +GF +++ S + +DG+ LGC + VS
Sbjct: 113 MEFESEDAAKIFYDEYARRLGF-VMRVMSCRRPERDGRILARRLGCNKEGYCVSIRGKFA 289
Query: 55 NLLKPNPTIRAQCMARVNMSMSLDGKITITKVALEHNHGL--SPTKSR 100
++ KP + R C A +++ + GK ITK +HNH L SP ++R
Sbjct: 290 SVRKPRASTREGCKAMIHIKYNKSGKWVITKFVKDHNHPLVVSPREAR 433
>TC232091
Length = 530
Score = 44.7 bits (104), Expect = 1e-04
Identities = 31/107 (28%), Positives = 52/107 (47%), Gaps = 6/107 (5%)
Frame = +1
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYFT---LGCTRARSYV---SNSK 54
M F SEE +Y+ YAR GF V+++ + + K C + +V + +K
Sbjct: 148 MEFQSEEAAKNFYEEYARREGF-VVRLDRCHRSEVDKQIISRRFSCNKQGFHVRVRNKTK 324
Query: 55 NLLKPNPTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRY 101
+ KP +IR C A + + ++ GK +TK EH+H L+ + Y
Sbjct: 325 PVHKPRASIREGCEAMMYVKVNTCGKWVVTKFVKEHSHLLNASALPY 465
>AW780793
Length = 419
Score = 42.4 bits (98), Expect = 7e-04
Identities = 29/93 (31%), Positives = 47/93 (50%), Gaps = 8/93 (8%)
Frame = +3
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYFTLG----CT----RARSYVSN 52
M F+S EE ++Y +Y R +GF TV+I+ N + +G C+ RA+ Y+
Sbjct: 144 MEFNSREEAREFYIAYGRRIGF-TVRIHH-NRRSRVNNQVIGQDFVCSKEGFRAKKYLHR 317
Query: 53 SKNLLKPNPTIRAQCMARVNMSMSLDGKITITK 85
+L P P R C A + +++ GK +TK
Sbjct: 318 KDRVLPPPPATREGCQAMIRLALRDRGKWVVTK 416
>AW099292
Length = 420
Score = 38.9 bits (89), Expect = 0.008
Identities = 15/38 (39%), Positives = 24/38 (62%)
Frame = +2
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKY 38
M F S E +YK YA+ GFGT K++S+ ++ K++
Sbjct: 272 MEFESHEAAYAFYKEYAKSAGFGTAKLSSRRSRASKEF 385
>BM525744
Length = 411
Score = 38.5 bits (88), Expect = 0.010
Identities = 17/49 (34%), Positives = 23/49 (46%)
Frame = +3
Query: 190 VDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGV 238
VDD +R++FW F V+ D TY N+Y +P VGV
Sbjct: 207 VDDSDAIRDIFWAHPDAIKLLGSFHTVLFLDNTYKVNRYQLPLLEIVGV 353
>TC234242
Length = 478
Score = 33.1 bits (74), Expect = 0.42
Identities = 16/65 (24%), Positives = 30/65 (45%)
Frame = +3
Query: 180 KNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGVN 239
+ Q+ + + D+ +R++FW V D+TY TN+Y +P FV ++
Sbjct: 42 ERDQYIHWHRIKDEDVVRDIFWCHPDSVKLVNACNLVFLIDSTYRTNRYRLPLLDFVVLD 221
Query: 240 HHGQS 244
G +
Sbjct: 222 FVGMT 236
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,211,625
Number of Sequences: 63676
Number of extensions: 445106
Number of successful extensions: 2490
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 2466
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2478
length of query: 694
length of database: 12,639,632
effective HSP length: 104
effective length of query: 590
effective length of database: 6,017,328
effective search space: 3550223520
effective search space used: 3550223520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC135795.1


