

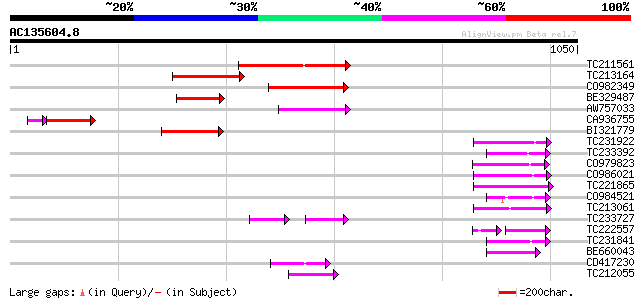
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135604.8 + phase: 0 /pseudo
(1050 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 197 3e-50
TC213164 139 9e-33
CO982349 125 1e-28
BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase... 125 1e-28
AW757033 108 9e-24
CA936755 93 8e-22
BI321779 97 3e-20
TC231922 83 7e-16
TC233392 82 9e-16
CO979823 78 2e-14
CO986021 76 9e-14
TC221865 75 2e-13
CO984521 73 6e-13
TC213061 72 2e-12
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 63 2e-12
TC222557 58 1e-11
TC231841 68 2e-11
BE660043 67 3e-11
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 67 5e-11
TC212055 66 7e-11
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 197 bits (500), Expect = 3e-50
Identities = 103/207 (49%), Positives = 136/207 (64%)
Frame = +1
Query: 424 LDGILIAN*VVDEARKLKKYLLLFKVDFAKAYDSVDWGYLDEVMGSMSFPVVWRKWIKEC 483
L LIAN V+DEA++ K L+FKVD+ KAYDSV W +L +M M F W KWI+EC
Sbjct: 1 LHSALIANEVIDEAKRSNKSCLVFKVDYEKAYDSVSWNFLMYMMWRMDFSPKWIKWIEEC 180
Query: 484 VGTATASVLVNGSPTDEFHLERGLRQGDPLSPFLFLLAA*GLNVLMKALVDAGMFTGYRV 543
V +A+ SVLVNGSPT EF +RGLRQGDPL+PFLF + A GLN LM+ V+ ++ Y V
Sbjct: 181 VKSASISVLVNGSPTAEFLPQRGLRQGDPLTPFLFNIVAEGLNGLMRRAVEENLYKPYLV 360
Query: 544 GRADSVVVSHLQFADDTLLIGNKSWANVRALRAGLVMFEAMSGLKVNFHKSSLVGVNIND 603
G A+ V +S LQ+ADDT+ G + NV A++ L FE +S L++NF KS + D
Sbjct: 361 G-ANGVPISILQYADDTIFFGEAAKENVEAIKVILRSFELVSNLRINFAKSCFGVFGVTD 537
Query: 604 SWLSEAASVLGCKVVKIPFLYLGLSRG 630
W EAA+ L C ++ PF+YLG+ G
Sbjct: 538 QWKQEAANYLHCSLLAFPFIYLGIPIG 618
>TC213164
Length = 446
Score = 139 bits (349), Expect = 9e-33
Identities = 63/135 (46%), Positives = 96/135 (70%)
Frame = +3
Query: 301 LTKPFSEDEIKAAVWDCDSFKSPGPDGINLGFFKSFWPEIKTDLVRFVTEFHRNGKLAKG 360
L F + E++A +WD + KSPGPDG+N F K FW +KTDL+RF+ EF+ NG K
Sbjct: 18 LVARFEKKEVRATMWDRGNAKSPGPDGLNFKFIKEFWDILKTDLLRFLDEFYANGVFLKR 197
Query: 361 INSTFIALIPKVASPQSLNEFRPISLVGSMYKILAKILANRLRQVIGSVVSEAQSAFVKN 420
N+ F+ALIPKV P SLNE++PISL+G +YKI+AK+L+ R ++V+ +++ E Q+ F++
Sbjct: 198 SNAFFLALIPKVHDP*SLNEYKPISLIGCIYKIVAKLLSGRHKKVLPTIIDERQTVFMEG 377
Query: 421 RQILDGILIAN*VVD 435
R L ++IAN +++
Sbjct: 378 RHTLHNVVIANEIME 422
>CO982349
Length = 795
Score = 125 bits (313), Expect = 1e-28
Identities = 65/149 (43%), Positives = 92/149 (61%)
Frame = +3
Query: 479 WIKECVGTATASVLVNGSPTDEFHLERGLRQGDPLSPFLFLLAA*GLNVLMKALVDAGMF 538
WI+ C+ +A+ S+LVN SP +EF +RGLRQGDPL+P LF + A GL LM+ VD F
Sbjct: 69 WIRGCLHSASVSILVNESPINEFSPQRGLRQGDPLAPLLFNIVAEGLTGLMREAVDRKRF 248
Query: 539 TGYRVGRADSVVVSHLQFADDTLLIGNKSWANVRALRAGLVMFEAMSGLKVNFHKSSLVG 598
+ VG+ + VS LQ+ADDT+ + N++ ++ L FE SGLK+NF +S
Sbjct: 249 NSFLVGK-NKEPVSILQYADDTIFFEEATMENMKVIKTILRCFELASGLKINFAESRFGA 425
Query: 599 VNINDSWLSEAASVLGCKVVKIPFLYLGL 627
+ D W EAA L C ++ +PF YLG+
Sbjct: 426 IWKPDHWCKEAAEFLNCSMLSMPFSYLGI 512
>BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (15%)
Length = 376
Score = 125 bits (313), Expect = 1e-28
Identities = 56/88 (63%), Positives = 69/88 (77%)
Frame = -2
Query: 310 IKAAVWDCDSFKSPGPDGINLGFFKSFWPEIKTDLVRFVTEFHRNGKLAKGINSTFIALI 369
IK+AVW C S KSPGPDG+N F K FW +K D++RF+ EFH NG KG N++FIALI
Sbjct: 375 IKSAVWQCGSVKSPGPDGLNFKFIKHFWERLKPDIIRFLNEFHANGIFPKGGNASFIALI 196
Query: 370 PKVASPQSLNEFRPISLVGSMYKILAKI 397
PKV PQ+LN+FRPISL+G +YKI+AKI
Sbjct: 195 PKVKHPQALNDFRPISLIGCVYKIVAKI 112
>AW757033
Length = 441
Score = 108 bits (271), Expect = 9e-24
Identities = 59/133 (44%), Positives = 79/133 (59%)
Frame = -2
Query: 498 TDEFHLERGLRQGDPLSPFLFLLAA*GLNVLMKALVDAGMFTGYRVGRADSVVVSHLQFA 557
T EF RGLRQGDPL+P LF +AA GL LM+ V + + VG+ V S LQ+A
Sbjct: 440 TREFSPHRGLRQGDPLAPLLFNIAAEGLTGLMREAVARNHYKSFAVGKYKKPV-SILQYA 264
Query: 558 DDTLLIGNKSWANVRALRAGLVMFEAMSGLKVNFHKSSLVGVNINDSWLSEAASVLGCKV 617
DDT+ G + NVR ++ L FE SGLK+NF KS +++ D W EAA + C +
Sbjct: 263 DDTIFFGEATMENVRVIKTILRGFELASGLKINFAKSRFGAISVPD*WRKEAAEFMNCSL 84
Query: 618 VKIPFLYLGLSRG 630
+ +PF YLG+ G
Sbjct: 83 LSLPFSYLGIPIG 45
>CA936755
Length = 424
Score = 93.2 bits (230), Expect(2) = 8e-22
Identities = 37/90 (41%), Positives = 56/90 (62%)
Frame = -2
Query: 69 FLLSVEWCMVWPNCIQVAQLRGLSDHCPILLSVDEENWGPRPVRLLKCWQDMPGYIDFIR 128
FL+S +W + WP+ Q R SDHCPI++ + +WGP+P R++ W GY +R
Sbjct: 294 FLVSEQWLLSWPDSSQYTLPRDFSDHCPIIMQTKKVDWGPKPFRVVDWWLHQKGYQRLVR 115
Query: 129 EKWVFFQVSGWGGHVLKEKLKLMKVALKDW 158
E W Q GWGG +LK KL+++K+++K W
Sbjct: 114 ETWSAEQQPGWGGILLKNKLRMLKLSIKQW 25
Score = 30.0 bits (66), Expect(2) = 8e-22
Identities = 13/36 (36%), Positives = 20/36 (55%)
Frame = -1
Query: 34 FNDFIEDRVLIDLLLSGRKFTWFKGDGRSMRRLDRF 69
FN +I + L ++ G +TW + + R RLDRF
Sbjct: 400 FNTWISEMELQEIKCVGSTYTWIRPNARVKSRLDRF 293
>BI321779
Length = 421
Score = 97.4 bits (241), Expect = 3e-20
Identities = 49/115 (42%), Positives = 70/115 (60%)
Frame = +2
Query: 282 RPGVGHLQFRKLSYLERGGLTKPFSEDEIKAAVWDCDSFKSPGPDGINLGFFKSFWPEIK 341
RP + ++F S + L + F +EIK + DC+S KSPGPDG +L K FW IK
Sbjct: 26 RPKLNGVEFLSFSNEDNAMLIQDFEVEEIKKVI*DCESSKSPGPDGYSLLLIKIFWEVIK 205
Query: 342 TDLVRFVTEFHRNGKLAKGINSTFIALIPKVASPQSLNEFRPISLVGSMYKILAK 396
+++ F EFH+ L +G N +FIALI K +P + ++ PISLVG +YKI+ K
Sbjct: 206 EEVINFFKEFHKFDFLPRGTNRSFIALIAKCDNPLDIGQYCPISLVGCLYKIV*K 370
>TC231922
Length = 803
Score = 82.8 bits (203), Expect = 7e-16
Identities = 42/145 (28%), Positives = 70/145 (47%), Gaps = 1/145 (0%)
Frame = -3
Query: 859 RWRRRLWAREEELVEECRALLLTVSLQESVTDRWLWVPNHDDGYSVRGVYDMLTSQEQPQ 918
+WRRRL+ E + + + +S+Q D W + YS + Y +L P
Sbjct: 537 KWRRRLFDYEYAMAVDFMNEISGISIQNQAHDSMFWKADSSGVYSTKSAYRLLMPSISPA 358
Query: 919 LHHNM-ELIWHK*VPLKVSILAWRLLRDRLPTKSNLANRGILALEARLCVSGCGNVEDVN 977
+ +++WH +P + ++ +WRL DRLPT+ NL+ R I + + GC + E+
Sbjct: 357 PSRRIFQILWHLKIPPRAAVFSWRLFLDRLPTRGNLSRRSIPIQDIMCPLCGCQH-EEAG 181
Query: 978 HLFLSCVTFSALWPMVRAWLGVVGS 1002
HLF C LW W+ V+G+
Sbjct: 180 HLFFHCKMTKGLWWESMRWIRVIGA 106
>TC233392
Length = 1145
Score = 82.4 bits (202), Expect = 9e-16
Identities = 45/119 (37%), Positives = 63/119 (52%), Gaps = 1/119 (0%)
Frame = -1
Query: 884 LQESVTDRWLWVPNHDDGYSVRGVYDMLTSQ-EQPQLHHNMELIWHK*VPLKVSILAWRL 942
+Q+ V D W+W ++ YS R Y +L E ++ +W +P K SI AWRL
Sbjct: 674 IQQQVPDTWVWKHEPNELYSTRSAYKLLQGHLEGEDQDGALQDLWKLKIPAKASIFAWRL 495
Query: 943 LRDRLPTKSNLANRGILALEARLCVSGCGNVEDVNHLFLSCVTFSALWPMVRAWLGVVG 1001
+RDRLPTKSNL R ++ LE LC ED +H+FL C LW + W+ +G
Sbjct: 494 IRDRLPTKSNLHRRQVV-LEDSLCPFCRIREEDASHIFLECNKIRPLWWESQTWVRSLG 321
>CO979823
Length = 853
Score = 78.2 bits (191), Expect = 2e-14
Identities = 43/144 (29%), Positives = 68/144 (46%), Gaps = 2/144 (1%)
Frame = -2
Query: 858 LRWRRRLWAREEELVEECRALLLTVSLQESVTDRWLWVPNHDDGYSVRGVYDMLTSQEQP 917
L WRR E + + + + D+WLW P YS + Y +L +
Sbjct: 798 LNWRRPXXDSEIAMADSFLGEITQQQIHPQREDKWLWKPEPGGHYSTKSGYHVLWGELTE 619
Query: 918 QLHH-NMELIWHK*VPLKVSILAWRLLRDRLPTKSNLANRGILALEARLCVSGCGNVED- 975
++ + IW +P K ++ AWRL+RDRLPTKSNL R ++ + + C N+E+
Sbjct: 618 EIQDADFAEIWKLKIPTKAAVFAWRLVRDRLPTKSNLRRRQVMVQD--MVCPLCNNIEEG 445
Query: 976 VNHLFLSCVTFSALWPMVRAWLGV 999
HLF +C LW +W+ +
Sbjct: 444 AAHLFFNCTKTLPLWWESMSWVNL 373
>CO986021
Length = 900
Score = 75.9 bits (185), Expect = 9e-14
Identities = 44/146 (30%), Positives = 70/146 (47%), Gaps = 2/146 (1%)
Frame = -1
Query: 859 RWRRRLWAREEELVEECRALLLTVSLQESVTDRWLWVPNHDDGYSVRGVYDMLTSQEQPQ 918
+WRR L+ E E + +S+ + + D LW + YS + Y +L +P
Sbjct: 876 KWRRNLFDHENEQAIAFMDDISAISIHQQLQDSMLWKADPTGIYSTKSAYRLLLPNNRPG 697
Query: 919 LHH-NMELIWHK*VPLKVSILAWRLLRDRLPTKSNLANRGILALEARLCVSGCGN-VEDV 976
H N +++W +P + + +WRL RDRL ++NL R + AL+ +C CGN E+
Sbjct: 696 QHSSNFKILWKLKIPPRAELFSWRLFRDRLSIRANLLRRHV-ALQDIMCPL-CGNHQEEA 523
Query: 977 NHLFLSCVTFSALWPMVRAWLGVVGS 1002
HLF C LW W +G+
Sbjct: 522 GHLFFHCRMTIGLWWESMNWTRTLGA 445
>TC221865
Length = 1045
Score = 74.7 bits (182), Expect = 2e-13
Identities = 46/150 (30%), Positives = 71/150 (46%), Gaps = 1/150 (0%)
Frame = -3
Query: 859 RWRRRLWAREEELVEECRALLLTVSLQESVTDRWLWVPNHDDGYSVRGVYDMLTSQE-QP 917
+WRR L+ E + + + +TD W+W + YS R Y ++ + +
Sbjct: 761 QWRRPLFDNEVDTTIGFLEDISRFPIHRHLTDCWVWKSEPNGHYSTRSAYHLMQHEAAKA 582
Query: 918 QLHHNMELIWHK*VPLKVSILAWRLLRDRLPTKSNLANRGILALEARLCVSGCGNVEDVN 977
E IW +P K +I A RL++DRLPTK+NL R + L LC+ E+ +
Sbjct: 581 NTDPAFEEIWKLKIPAKAAIFA*RLVKDRLPTKNNLRRRQV-QLNGTLCLFCRNYEEEAS 405
Query: 978 HLFLSCVTFSALWPMVRAWLGVVGSRFSFN 1007
HLF SC +W +W+ G+ FS N
Sbjct: 404 HLFFSCTKTQPVWWESLSWINTSGA-FSAN 318
>CO984521
Length = 716
Score = 73.2 bits (178), Expect = 6e-13
Identities = 42/124 (33%), Positives = 68/124 (53%), Gaps = 5/124 (4%)
Frame = -2
Query: 883 SLQESVTDRWLWVPNHDDGYSVRGVYDML----TSQEQPQLHHNMELIWHK*VPLKVSIL 938
+++ ++D+W W YS + Y L +EQ + +W VPLKV+I
Sbjct: 667 TIRPELSDQWKWAAEPSGCYSTKSAYKALHHVTVGEEQDG---KFKELWKLRVPLKVAIF 497
Query: 939 AWRLLRDRLPTKSNLANRGILALEARLCVSGCGNVED-VNHLFLSCVTFSALWPMVRAWL 997
AWRL++D+LPTK+NL + + L+ LC C +VE+ +HLF C S LW ++W+
Sbjct: 496 AWRLIQDKLPTKANLRKKRV-ELQEYLCPL-CRSVEETASHLFFHCSKVSPLWWESQSWV 323
Query: 998 GVVG 1001
++G
Sbjct: 322 NMMG 311
>TC213061
Length = 823
Score = 71.6 bits (174), Expect = 2e-12
Identities = 43/146 (29%), Positives = 70/146 (47%), Gaps = 2/146 (1%)
Frame = +2
Query: 859 RWRRRLWAREEELVEECRALLLTVSLQESVTDRWLWVPNHDDGYSVRGVYDMLTS--QEQ 916
+WRR L+ E E++ + ++ + D+W+W Y R Y ++ E+
Sbjct: 35 QWRRPLFDSEIEMLVAFIQEVEGHKIRSDIEDQWVWAAESSGSYLARSAYRVIREGIPEE 214
Query: 917 PQLHHNMELIWHK*VPLKVSILAWRLLRDRLPTKSNLANRGILALEARLCVSGCGNVEDV 976
Q EL W VP+KV++ AWRLL+D LPT+ NL + + L +C E
Sbjct: 215 EQDREFKEL-WKLKVPMKVTMFAWRLLKDILPTRDNLRRKRV-ELHEYVCPLCRSMDESA 388
Query: 977 NHLFLSCVTFSALWPMVRAWLGVVGS 1002
+HLF C +W +W+ +VG+
Sbjct: 389 SHLFFHCSKILPIWWESLSWVKLVGA 466
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 62.8 bits (151), Expect(2) = 2e-12
Identities = 29/74 (39%), Positives = 44/74 (59%)
Frame = -3
Query: 445 LLFKVDFAKAYDSVDWGYLDEVMGSMSFPVVWRKWIKECVGTATASVLVNGSPTDEFHLE 504
L K+D KA+D++D +L V+ + + WI+ + +A S+ VNG P F +
Sbjct: 765 LALKIDIVKAFDTLDRDFLISVLNKFGYSHTFCNWIRVILLSAKLSISVNGEPVGFFSCQ 586
Query: 505 RGLRQGDPLSPFLF 518
RG+RQGDPLSP L+
Sbjct: 585 RGVRQGDPLSPSLY 544
Score = 28.5 bits (62), Expect(2) = 2e-12
Identities = 21/80 (26%), Positives = 36/80 (44%)
Frame = -2
Query: 548 SVVVSHLQFADDTLLIGNKSWANVRALRAGLVMFEAMSGLKVNFHKSSLVGVNINDSWLS 607
S+ SH+ ADD ++ + NV L +++ G ++ HKS + +I L
Sbjct: 463 SLTRSHVMHADDIMIFCKGTSRNVHNLINFFQLYKDSFGQVISKHKSKIFIGSIPGQRLR 284
Query: 608 EAASVLGCKVVKIPFLYLGL 627
+ +L IPF Y G+
Sbjct: 283 VISDLLQYFHSDIPFNYFGV 224
>TC222557
Length = 1002
Score = 58.2 bits (139), Expect(2) = 1e-11
Identities = 30/84 (35%), Positives = 44/84 (51%)
Frame = +1
Query: 918 QLHHNMELIWHK*VPLKVSILAWRLLRDRLPTKSNLANRGILALEARLCVSGCGNVEDVN 977
+L +E +W +PLK + AWRL+++R+PTK NL R + L +C E+ +
Sbjct: 415 RLDEALEDLWQLKIPLKATTFAWRLIKERIPTKGNLWRRRV-QLNNLMCPFCNRQEEEAS 591
Query: 978 HLFLSCVTFSALWPMVRAWLGVVG 1001
HLF +C LW AW VG
Sbjct: 592 HLFFNCPRILPLWWESLAWTKTVG 663
Score = 30.4 bits (67), Expect(2) = 1e-11
Identities = 18/57 (31%), Positives = 26/57 (45%), Gaps = 3/57 (5%)
Frame = +3
Query: 858 LRWRRRLWAREEELVEECRALLLTVS---LQESVTDRWLWVPNHDDGYSVRGVYDML 911
L WRR L+ E V+ + L +S + D W+W P + +S R Y ML
Sbjct: 234 LEWRRHLFDNE---VQAAASFLDDISWGHIDRWTLDCWVWKPEPNGQFSTRSAYRML 395
>TC231841
Length = 791
Score = 68.2 bits (165), Expect = 2e-11
Identities = 37/119 (31%), Positives = 60/119 (50%), Gaps = 1/119 (0%)
Frame = +3
Query: 884 LQESVTDRWLWVPNHDDGYSVRGVYDMLTSQE-QPQLHHNMELIWHK*VPLKVSILAWRL 942
+Q D+W+W + Y+ + Y +L + + Q E +W +P K++I AWRL
Sbjct: 45 IQPQQGDQWVWKADPSGQYTAKSAYGVLWGEMFEEQQDGVFEELWKLKLPSKITIFAWRL 224
Query: 943 LRDRLPTKSNLANRGILALEARLCVSGCGNVEDVNHLFLSCVTFSALWPMVRAWLGVVG 1001
+RDRLPT+SNL + I + R C E HLF C + +W +W+ ++G
Sbjct: 225 IRDRLPTRSNLRRKQIEVDDPR-CPFCRSAEESAAHLFFHCSRIAPVWWESLSWVNLLG 398
>BE660043
Length = 714
Score = 67.4 bits (163), Expect = 3e-11
Identities = 38/102 (37%), Positives = 53/102 (51%), Gaps = 1/102 (0%)
Frame = -2
Query: 883 SLQESVTDRWLWVPNHDDGYSVRGVYDMLT-SQEQPQLHHNMELIWHK*VPLKVSILAWR 941
+LQ+ D W+W P + YS R Y +L + E+ + + +W +P K SI A R
Sbjct: 647 ALQQHAADSWVWKPESNGYYSSRNAYILLQGNSEEGNMDDIFKDLWKLKIPAKASIFAGR 468
Query: 942 LLRDRLPTKSNLANRGILALEARLCVSGCGNVEDVNHLFLSC 983
L+RDRLPTKSN+ R I + LC E +HLF C
Sbjct: 467 LIRDRLPTKSNMRKRQI-DINDSLCPFCSIKEETASHLFFDC 345
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like
protein {Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 66.6 bits (161), Expect = 5e-11
Identities = 38/112 (33%), Positives = 60/112 (52%)
Frame = -3
Query: 483 CVGTATASVLVNGSPTDEFHLERGLRQGDPLSPFLFLLAA*GLNVLMKALVDAGMFTGYR 542
C+ T T S+L NG D F G+RQ DP++P+LF+L L+ L+ +++ + +
Sbjct: 668 CMSTTTMSLLWNGEVLDHFSPNIGVRQRDPIAPYLFVLCLERLSHLINLVMNKKL*RSIQ 489
Query: 543 VGRADSVVVSHLQFADDTLLIGNKSWANVRALRAGLVMFEAMSGLKVNFHKS 594
V R +++SHL F DD +L + V + L +F SG KVN K+
Sbjct: 488 VNRG-GLLISHLAFVDDLILFVEANMDQVDVINLTLELFSDSSGAKVNVDKT 336
>TC212055
Length = 776
Score = 66.2 bits (160), Expect = 7e-11
Identities = 35/93 (37%), Positives = 56/93 (59%)
Frame = +1
Query: 517 LFLLAA*GLNVLMKALVDAGMFTGYRVGRADSVVVSHLQFADDTLLIGNKSWANVRALRA 576
LF + A GL LM+ +D +++ VG+ + + V+ LQ+ADDT+ +G + NV +++
Sbjct: 1 LFNIVAEGLTGLMREALDKSLYSSLMVGK-NKIPVNILQYADDTIFLGEATMQNVMTIKS 177
Query: 577 GLVMFEAMSGLKVNFHKSSLVGVNINDSWLSEA 609
L +FE SGLK++F KSS + D W EA
Sbjct: 178 MLRVFELASGLKIHFAKSSFGAIGKPDHWQKEA 276
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.338 0.149 0.505
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 53,106,904
Number of Sequences: 63676
Number of extensions: 849595
Number of successful extensions: 6271
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 6108
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6224
length of query: 1050
length of database: 12,639,632
effective HSP length: 107
effective length of query: 943
effective length of database: 5,826,300
effective search space: 5494200900
effective search space used: 5494200900
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC135604.8


