

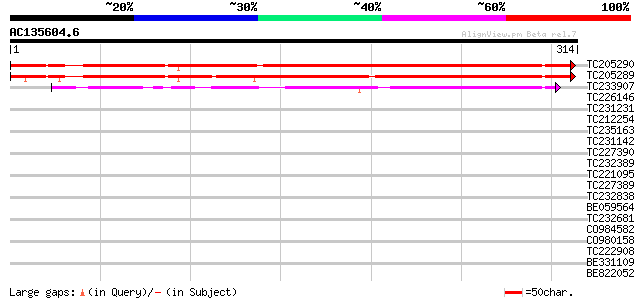
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135604.6 - phase: 0
(314 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC205290 399 e-112
TC205289 similar to UP|Q8V7E9 (Q8V7E9) ORF1 (Fragment), partial ... 376 e-105
TC233907 similar to UP|Q86NQ6 (Q86NQ6) RE01075p, partial (10%) 160 5e-40
TC226146 weakly similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine p... 40 0.002
TC231231 weakly similar to UP|Q88TJ4 (Q88TJ4) Cell surface prote... 35 0.044
TC212254 similar to UP|Q9FMJ0 (Q9FMJ0) Phosphatidylinositol 4-ki... 33 0.17
TC235163 similar to UP|Q9XYL1 (Q9XYL1) Development protein DG110... 33 0.17
TC231142 weakly similar to UP|Q7PC53 (Q7PC53) Chitinase B, parti... 33 0.22
TC227390 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacety... 32 0.29
TC232389 31 0.64
TC221095 31 0.64
TC227389 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacety... 31 0.64
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 31 0.83
BE059564 30 1.1
TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g4... 30 1.1
CO984582 30 1.1
CO980158 30 1.1
TC222908 30 1.1
BE331109 30 1.4
BE822052 similar to GP|4557063|gb|A expressed protein {Arabidops... 30 1.4
>TC205290
Length = 1165
Score = 399 bits (1026), Expect = e-112
Identities = 215/317 (67%), Positives = 249/317 (77%), Gaps = 4/317 (1%)
Frame = +3
Query: 1 MQVPLIDRLNDFQAGFNSLQQNPSFPSQITATSFNGIQSVSIAFNFCKWGAVILALVATF 60
MQVPLIDR+ND Q G NSLQ NPSFPSQIT S+A+NFCKWGAVILALVATF
Sbjct: 78 MQVPLIDRINDLQVGLNSLQ-NPSFPSQITT---------SLAYNFCKWGAVILALVATF 227
Query: 61 TSLINKVTIFIIHLRKKASSLPSITFDNDDDF--SSDDEDN-ENDDIVSLSSSSEFEDDE 117
S+IN+VTIFIIH R KA SLPS+ NDDDF SSDDED+ +ND+ L+ SS +DE
Sbjct: 228 GSIINRVTIFIIHFRAKAPSLPSLNL-NDDDFYTSSDDEDDFQNDEDDDLTYSSSEPEDE 404
Query: 118 PSMSSSS-SFKDFFRLTGNSNDYDVNNVFQTQNSGHRRQHSIGDFFSLLELANGDNVVKL 176
PS SSS +D+FR+ G N +++ FQTQN H+R SIGD FSL ELAN +VVKL
Sbjct: 405 PSASSSFIRLEDYFRIRGTDN---LDDEFQTQNGTHKRCRSIGDIFSLSELANSKSVVKL 575
Query: 177 WDSIGFGLGLDFDEYEDGVISSTNPHAPLTSAAASPDVIVSAGEGAHGNLAVEIWDTRLR 236
WDSIGFGLGLDFD+Y+ +SS P TS+ ASP V+VSAGEGA+GNLA+EIWDTRLR
Sbjct: 576 WDSIGFGLGLDFDDYDGYEVSSAKPLQAPTSSTASPAVVVSAGEGAYGNLALEIWDTRLR 755
Query: 237 RRKPSVVAEWGPTVGNTLRVESGGVQKVYVRDNGRQRLTVGDMRKVSFPLGNVTESDADN 296
RRKP+VVAEWGPTVG TL VESGGVQKVYVRD+GR +TVGDMRKVS PLGNVTESDAD
Sbjct: 756 RRKPTVVAEWGPTVGKTLHVESGGVQKVYVRDDGRYSVTVGDMRKVSTPLGNVTESDAD- 932
Query: 297 TWWDADAVIVTDESYGK 313
TWWDADA++V+DES+G+
Sbjct: 933 TWWDADAIVVSDESFGE 983
>TC205289 similar to UP|Q8V7E9 (Q8V7E9) ORF1 (Fragment), partial (48%)
Length = 1305
Score = 376 bits (966), Expect = e-105
Identities = 211/325 (64%), Positives = 250/325 (76%), Gaps = 12/325 (3%)
Frame = +3
Query: 1 MQVPLID--RLNDFQAGFNSLQQNPSFP--SQITATSFNGIQSVSIAFNFCKWGAVILAL 56
MQVPLID R+ND Q G NSLQ NPSFP SQIT S+A+NFCKWGAVILAL
Sbjct: 51 MQVPLIDIDRINDLQVGLNSLQ-NPSFPIPSQITT---------SLAYNFCKWGAVILAL 200
Query: 57 VATFTSLINKVTIFIIHLRKKASSLPSITFDNDDDF-----SSDDEDNENDDIVSLSSSS 111
VATF S+IN+VTIFII R KA SLPS+T NDD+F S DD+D++NDD ++ SSS
Sbjct: 201 VATFGSIINRVTIFIIRFRAKAPSLPSLTL-NDDNFYTSSSSDDDDDDDNDDDLTYSSSE 377
Query: 112 EFEDDEPSMSSSS-SFKDFFRLTG--NSNDYDVNNVFQTQNSGHRRQHSIGDFFSLLELA 168
+DEPS SSS D+FRL G N+N+ ++++ FQTQN H+R SIGD FSL ELA
Sbjct: 378 P--EDEPSASSSFIRLDDYFRLRGTDNNNNNNLDDEFQTQNGTHKRCRSIGDIFSLSELA 551
Query: 169 NGDNVVKLWDSIGFGLGLDFDEYEDGVISSTNPHAPLTSAAASPDVIVSAGEGAHGNLAV 228
N +VVKLWD+IGFGLGLDFD+Y+ + + A TS+ ASP V+VSAGEGA GNLA+
Sbjct: 552 NSKSVVKLWDTIGFGLGLDFDDYDGYYVGNA---AVPTSSTASPSVVVSAGEGACGNLAL 722
Query: 229 EIWDTRLRRRKPSVVAEWGPTVGNTLRVESGGVQKVYVRDNGRQRLTVGDMRKVSFPLGN 288
EIWDTRLRRRKP+VVAEWGPTVG TLRVESGGVQKVYVRD+GR +TVGDMRKVS PL +
Sbjct: 723 EIWDTRLRRRKPTVVAEWGPTVGKTLRVESGGVQKVYVRDDGRYGVTVGDMRKVSTPLES 902
Query: 289 VTESDADNTWWDADAVIVTDESYGK 313
VTESDAD TWWDADA++V+DES+G+
Sbjct: 903 VTESDAD-TWWDADAIVVSDESFGE 974
>TC233907 similar to UP|Q86NQ6 (Q86NQ6) RE01075p, partial (10%)
Length = 845
Score = 160 bits (406), Expect = 5e-40
Identities = 104/287 (36%), Positives = 152/287 (52%), Gaps = 5/287 (1%)
Frame = +1
Query: 24 SFPSQITATSFNGIQSVSIAFNFCKWGAVILALVATFTSLINKVTIFIIHLRKKASSLPS 83
SF +T S NG+ F+F + + + + + T L ++ I+ + + P+
Sbjct: 43 SFLPLLTLLSANGV------FSFSAYSSPLSSSASAKTQLPFLLSSLIVII-----ATPT 189
Query: 84 ITFDNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSSFKDFFRLTGNSNDYDVNN 143
+ D+ S E ++DD E E++E ++FR+ G++ND
Sbjct: 190 LY----DEIYSTSEFEDDDD--------EEEEEEEEEEEEERRDEYFRVRGSTND----- 318
Query: 144 VFQTQNSGHRRQHSIGDFFSLLELANGDNVVKLWDSIGFGLGLDFDEYE-----DGVISS 198
R SIGDF SL E+AN +VVKLWD+IGFGLG F+ ++ V+S
Sbjct: 319 ---------ERCRSIGDFLSLSEIANTKSVVKLWDTIGFGLGFGFEHFDRSSEGSSVVSV 471
Query: 199 TNPHAPLTSAAASPDVIVSAGEGAHGNLAVEIWDTRLRRRKPSVVAEWGPTVGNTLRVES 258
+ P P V+VSAG+ A GNLAV IWDTRLRRR P+++AEWGP++G T+ VES
Sbjct: 472 YDGELP------DPAVVVSAGQNASGNLAVRIWDTRLRRRIPAMMAEWGPSLGKTVGVES 633
Query: 259 GGVQKVYVRDNGRQRLTVGDMRKVSFPLGNVTESDADNTWWDADAVI 305
V KV+V+D+GR TVG+MR PL VT+S D WW ++
Sbjct: 634 SEVHKVFVKDDGRYEFTVGNMRNAMSPLQYVTDSHLD-LWWPNSLIV 771
>TC226146 weakly similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein
kinase-like protein (CBL-interacting protein kinase 5),
partial (37%)
Length = 1110
Score = 39.7 bits (91), Expect = 0.002
Identities = 22/75 (29%), Positives = 42/75 (55%)
Frame = +2
Query: 74 LRKKASSLPSITFDNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSSFKDFFRLT 133
+R A S+ +++ DFS DD N +DD S SS+++ + + + S+ F ++
Sbjct: 269 MRPIAFSMKESAVEDNIDFSGDDVSNGSDDGNSSSSNNDDGELTGAKPARPSYNAFEIIS 448
Query: 134 GNSNDYDVNNVFQTQ 148
SN +D+ N+F+T+
Sbjct: 449 SLSNGFDLRNLFETR 493
>TC231231 weakly similar to UP|Q88TJ4 (Q88TJ4) Cell surface protein, partial
(10%)
Length = 1077
Score = 35.0 bits (79), Expect = 0.044
Identities = 22/63 (34%), Positives = 35/63 (54%)
Frame = +3
Query: 76 KKASSLPSITFDNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSSFKDFFRLTGN 135
KK S++ S+ D+ +SS +D+ +DD++S SSS+ E S S SS D +
Sbjct: 504 KKTSTILSLPKDSKC-YSSSGDDSSSDDLLSDDSSSDDSSSEDSSSDDSSSDDLSSDDSS 680
Query: 136 SND 138
S+D
Sbjct: 681 SDD 689
Score = 32.0 bits (71), Expect = 0.37
Identities = 22/55 (40%), Positives = 31/55 (56%)
Frame = +3
Query: 74 LRKKASSLPSITFDNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSSFKD 128
L +SS S + D+ D SS D D +DD S SSS+ + +PS+S+ S KD
Sbjct: 588 LSDDSSSDDSSSEDSSSDDSSSD-DLSSDDSSSDDSSSDDDSSDPSISNESIKKD 749
>TC212254 similar to UP|Q9FMJ0 (Q9FMJ0) Phosphatidylinositol 4-kinase
(At5g64070/MHJ24_5), partial (12%)
Length = 683
Score = 33.1 bits (74), Expect = 0.17
Identities = 21/68 (30%), Positives = 33/68 (47%), Gaps = 3/68 (4%)
Frame = +2
Query: 91 DFSSDDEDNENDDIV-SLSSSSEFEDDEPSMSSSSSFKDFFRLTGNSND--YDVNNVFQT 147
D DDED+E D L S ED++ + SS FK FR + N ++ + ++
Sbjct: 467 DSRGDDEDSEKDGFFRRLLRDSRSEDEDVASSSEGLFKRLFRDSXNDSEDRTRTKTIEES 646
Query: 148 QNSGHRRQ 155
SG R++
Sbjct: 647 TTSGRRKE 670
>TC235163 similar to UP|Q9XYL1 (Q9XYL1) Development protein DG1106
(Fragment), partial (5%)
Length = 927
Score = 33.1 bits (74), Expect = 0.17
Identities = 23/52 (44%), Positives = 28/52 (53%)
Frame = +3
Query: 74 LRKKASSLPSITFDNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSS 125
L KK SS + D+DD FSS + + N D S SSS + D SSSSS
Sbjct: 486 LSKKLSSSCKCS-DSDDGFSSLNSYSRNSDDESTSSSPSSDSDSECCSSSSS 638
Score = 27.7 bits (60), Expect = 7.0
Identities = 34/172 (19%), Positives = 63/172 (35%), Gaps = 15/172 (8%)
Frame = +3
Query: 111 SEFEDDEPSMSSSSSFKDFFRLTGNSNDYDVNNVFQTQNSGHRRQHSIGDFFSL-----L 165
+ FEDD+ + +S + + +N+ + N+ N+ + Q ++ S L
Sbjct: 279 ARFEDDQSKSCNFTSANNILSINNCNNNINNNSNNTNNNNRNNNQINVNSNVSTTGGTPL 458
Query: 166 ELANGD---------NVVKLWDSI-GFGLGLDFDEYEDGVISSTNPHAPLTSAAASPDVI 215
+A GD + K DS GF + D +S++P + S S
Sbjct: 459 RIAEGDLSGLSKKLSSSCKCSDSDDGFSSLNSYSRNSDDESTSSSPSSDSDSECCSSSSS 638
Query: 216 VSAGEGAHGNLAVEIWDTRLRRRKPSVVAEWGPTVGNTLRVESGGVQKVYVR 267
S+ + R +PS+ + N LR+ G+Q + R
Sbjct: 639 TSSST-----------PNQSRENEPSIAESSPEYIVNLLRLHIQGIQSILTR 761
>TC231142 weakly similar to UP|Q7PC53 (Q7PC53) Chitinase B, partial (3%)
Length = 1130
Score = 32.7 bits (73), Expect = 0.22
Identities = 10/38 (26%), Positives = 25/38 (65%)
Frame = +1
Query: 88 NDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSS 125
+DDD + ++D++ND+++ + ++ E + + +MS S
Sbjct: 544 SDDDVEASEDDDDNDNVIGIENNGEEDSEADAMSEEGS 657
>TC227390 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (24%)
Length = 808
Score = 32.3 bits (72), Expect = 0.29
Identities = 15/31 (48%), Positives = 19/31 (60%)
Frame = +1
Query: 87 DNDDDFSSDDEDNENDDIVSLSSSSEFEDDE 117
D DDDF S DE+ E+ D S S + EDD+
Sbjct: 199 DEDDDFGSSDEEMEDADSDSDDESDDKEDDD 291
Score = 27.7 bits (60), Expect = 7.0
Identities = 16/42 (38%), Positives = 23/42 (54%)
Frame = +1
Query: 87 DNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSSFKD 128
DN++D SDD+ +E+DD SS E ED + S K+
Sbjct: 169 DNEED--SDDDSDEDDDFG--SSDEEMEDADSDSDDESDDKE 282
>TC232389
Length = 629
Score = 31.2 bits (69), Expect = 0.64
Identities = 18/45 (40%), Positives = 27/45 (60%), Gaps = 7/45 (15%)
Frame = +3
Query: 80 SLPSITFDNDDD-----FSSDDEDNENDDIV--SLSSSSEFEDDE 117
SLPS + +DD F D N+N+DI+ SL+S ++E+DE
Sbjct: 57 SLPSDSESSDDSIMGESFEMDHIGNQNEDIISESLNSDDDYEEDE 191
>TC221095
Length = 446
Score = 31.2 bits (69), Expect = 0.64
Identities = 17/46 (36%), Positives = 26/46 (55%), Gaps = 1/46 (2%)
Frame = +3
Query: 81 LPSITF-DNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSS 125
LPS T D+D D+ D+ ++ VSL S S+ E ++ S + SS
Sbjct: 51 LPSPTLSDSDSDYEDSSADDHHESSVSLDSQSQSELEDASPENGSS 188
>TC227389 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (63%)
Length = 1144
Score = 31.2 bits (69), Expect = 0.64
Identities = 17/42 (40%), Positives = 23/42 (54%)
Frame = +3
Query: 76 KKASSLPSITFDNDDDFSSDDEDNENDDIVSLSSSSEFEDDE 117
KK + S +DDDF S DE+ E+ D S S + +DDE
Sbjct: 534 KKDNEEDSDDDSDDDDFGSSDEEMEDADSDSDDESGDEDDDE 659
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 30.8 bits (68), Expect = 0.83
Identities = 13/31 (41%), Positives = 17/31 (53%)
Frame = +2
Query: 87 DNDDDFSSDDEDNENDDIVSLSSSSEFEDDE 117
++DDD DD D+E DD + EDDE
Sbjct: 353 EDDDDDEGDDNDDEEDDAPDGGDDDDDEDDE 445
Score = 28.9 bits (63), Expect = 3.2
Identities = 14/74 (18%), Positives = 33/74 (43%)
Frame = +2
Query: 87 DNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSSFKDFFRLTGNSNDYDVNNVFQ 146
D+DD+ S DD D++ D+ E +++E + R + + + ++ F+
Sbjct: 479 DDDDNDSDDDSDDDEDE-------DEEDEEEQGEEEDLGTEYLIRPLETAEEEEASSDFE 637
Query: 147 TQNSGHRRQHSIGD 160
+ +G + + D
Sbjct: 638 PEENGEEEEEDVDD 679
Score = 28.1 bits (61), Expect = 5.4
Identities = 17/66 (25%), Positives = 31/66 (46%), Gaps = 2/66 (3%)
Frame = +2
Query: 65 NKVTIFIIHLRKKASSLPSITFD--NDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSS 122
N+V + + ++ S+ + D +D+D DD+D+E DD + + EDD P
Sbjct: 260 NEVKVLVEEDDEEYESVDEVVDDAEDDEDEEEDDDDDEGDD------NDDEEDDAPDGGD 421
Query: 123 SSSFKD 128
+D
Sbjct: 422 DDDDED 439
>BE059564
Length = 406
Score = 30.4 bits (67), Expect = 1.1
Identities = 15/47 (31%), Positives = 27/47 (56%), Gaps = 5/47 (10%)
Frame = +3
Query: 84 ITFDNDDDFSSDDE-----DNENDDIVSLSSSSEFEDDEPSMSSSSS 125
++ D+DDD +DDE D+E+++ V + D P +S+S+S
Sbjct: 183 LSHDDDDDDETDDEEEDDDDDEDEEYVPAGAPPRVSHDAPRVSTSNS 323
>TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g47970
{Arabidopsis thaliana;} , partial (39%)
Length = 867
Score = 30.4 bits (67), Expect = 1.1
Identities = 13/34 (38%), Positives = 22/34 (64%)
Frame = +1
Query: 84 ITFDNDDDFSSDDEDNENDDIVSLSSSSEFEDDE 117
++FD+DDD + D+ED++ DD + ED+E
Sbjct: 202 LSFDDDDD-NEDEEDDDEDDAPDGGDDDDDEDEE 300
>CO984582
Length = 849
Score = 30.4 bits (67), Expect = 1.1
Identities = 18/88 (20%), Positives = 39/88 (43%), Gaps = 9/88 (10%)
Frame = -1
Query: 72 IHLRKKASSLPSITFDNDDDF---------SSDDEDNENDDIVSLSSSSEFEDDEPSMSS 122
I++ + S P + D ++D+ SDD + EN+ + ED+E S
Sbjct: 576 INIDDSSCSYPLVQVDEEEDYYDGTQHSDYESDDSNAENNPLNDYPDEVSEEDEEDENSE 397
Query: 123 SSSFKDFFRLTGNSNDYDVNNVFQTQNS 150
+ S ++ + S+D ++ + F ++
Sbjct: 396 NESEREDGNTSSESSDENIEHGFSEDDA 313
>CO980158
Length = 803
Score = 30.4 bits (67), Expect = 1.1
Identities = 18/88 (20%), Positives = 39/88 (43%), Gaps = 9/88 (10%)
Frame = -3
Query: 72 IHLRKKASSLPSITFDNDDDF---------SSDDEDNENDDIVSLSSSSEFEDDEPSMSS 122
I++ + S P + D ++D+ SDD + EN+ + ED+E S
Sbjct: 471 INIDDSSCSYPLVQVDEEEDYYDGTQHSDYESDDSNAENNPLNDYPDEVSEEDEEDENSE 292
Query: 123 SSSFKDFFRLTGNSNDYDVNNVFQTQNS 150
+ S ++ + S+D ++ + F ++
Sbjct: 291 NESEREDGNTSSESSDENIEHGFSEDDA 208
>TC222908
Length = 535
Score = 30.4 bits (67), Expect = 1.1
Identities = 14/26 (53%), Positives = 16/26 (60%)
Frame = +1
Query: 234 RLRRRKPSVVAEWGPTVGNTLRVESG 259
R R +KP+VVAEWGPT R G
Sbjct: 319 RNRSQKPTVVAEWGPTEDRRQRRRRG 396
>BE331109
Length = 475
Score = 30.0 bits (66), Expect = 1.4
Identities = 15/49 (30%), Positives = 23/49 (46%)
Frame = -1
Query: 42 IAFNFCKWGAVILALVATFTSLINKVTIFIIHLRKKASSLPSITFDNDD 90
+ F FC WG + L + TSL N + ++ L SS T+ + D
Sbjct: 211 VYFRFCLWGCLCLVKMIILTSLYNFFSSSVVLLSSS*SSFAPATWGSAD 65
>BE822052 similar to GP|4557063|gb|A expressed protein {Arabidopsis
thaliana}, partial (28%)
Length = 668
Score = 30.0 bits (66), Expect = 1.4
Identities = 13/31 (41%), Positives = 17/31 (53%)
Frame = -1
Query: 87 DNDDDFSSDDEDNENDDIVSLSSSSEFEDDE 117
D+DD DD+DN+ DD E ED+E
Sbjct: 374 DDDDSDEDDDDDNDGDDDGEDEDEKEGEDEE 282
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.133 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,759,981
Number of Sequences: 63676
Number of extensions: 171774
Number of successful extensions: 1771
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 1321
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1569
length of query: 314
length of database: 12,639,632
effective HSP length: 97
effective length of query: 217
effective length of database: 6,463,060
effective search space: 1402484020
effective search space used: 1402484020
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC135604.6


