

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135565.15 + phase: 0
(557 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
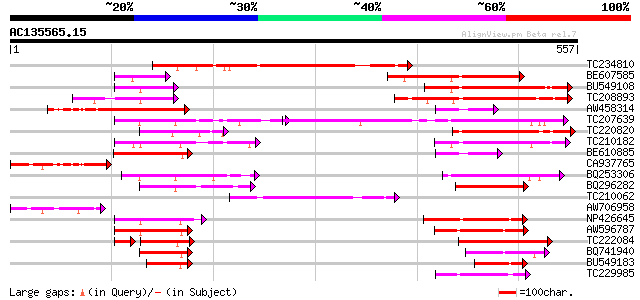
Score E
Sequences producing significant alignments: (bits) Value
TC234810 223 2e-58
BE607585 195 5e-50
BU549108 189 2e-48
TC208893 similar to UP|O48592 (O48592) GT2 protein, partial (11%) 187 1e-47
AW458314 183 2e-46
TC207639 similar to UP|O48592 (O48592) GT2 protein, partial (24%) 155 3e-38
TC220820 similar to UP|O48592 (O48592) GT2 protein, partial (11%) 155 4e-38
TC210182 similar to UP|Q9AVE4 (Q9AVE4) DNA-binding protein DF1, ... 120 2e-27
BE610885 119 3e-27
CA937765 117 1e-26
BQ253306 107 1e-23
BQ296282 homologue to GP|18182309|gb GT-2 factor {Glycine max}, ... 99 5e-21
TC210062 GT-2 factor [Glycine max] 88 1e-17
AW706958 homologue to GP|3056598|gb|A T1F9.19 {Arabidopsis thali... 87 3e-17
NP426645 GT-2 factor [Glycine max] 80 2e-15
AW596787 similar to GP|18182311|gb| GT-2 factor {Glycine max}, p... 78 9e-15
TC222084 similar to UP|O48592 (O48592) GT2 protein, partial (13%) 65 8e-11
BQ741940 homologue to GP|18182311|gb| GT-2 factor {Glycine max},... 62 5e-10
BU549183 similar to GP|18182311|gb GT-2 factor {Glycine max}, pa... 49 6e-06
TC229985 44 2e-04
>TC234810
Length = 747
Score = 223 bits (568), Expect = 2e-58
Identities = 132/269 (49%), Positives = 174/269 (64%), Gaps = 14/269 (5%)
Frame = +1
Query: 141 KRSAEKCKEKFEEESRFFNN-INH--NQNSFGKNFRFVTELEEVY--QGGGGENNKNLVE 195
+R EKCKEKFEEESR+FNN IN+ N N+ N+RF++ELE++Y QG G++ + + +
Sbjct: 1 EREREKCKEKFEEESRYFNNNINYAKNNNNSTSNYRFLSELEQLYHQQGSSGDHLEKMTQ 180
Query: 196 AEKQNEVQDKMDPH-------EEDSR--MDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDD 246
Q Q +MD H E DSR + D V+K +E + ++KRKRS D
Sbjct: 181 PPLQK--QGRMDHHALELEEEEGDSRNVIVDASVTKIQSDEALAVEKITKDRKRKRS--D 348
Query: 247 RFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKELELM 306
RFE+FKGFCES+V KMM QQEEMHNKL+EDM+KRDEEKF+REEAWKKQEMEKMNKELE+M
Sbjct: 349 RFEMFKGFCESIVHKMMTQQEEMHNKLLEDMMKRDEEKFTREEAWKKQEMEKMNKELEMM 528
Query: 307 AHEQAIAGDRQAHIIQFLNKFSTSANSSSLTSMSTQLQAYLATLTSNSSSSTLHSQNPNP 366
A EQA+AGDRQA IIQ LNKFS + +S + ++ +T SQNPNP
Sbjct: 529 AREQAVAGDRQAKIIQILNKFSATTSSPASHTL--------------KKVNTHISQNPNP 666
Query: 367 ETLKKTLQPIPENPSSTLPSSSTTLVAQP 395
+ + ++ + +PS+S+T P
Sbjct: 667 SQTENPTLSVAQD--TLIPSTSSTSTPAP 747
>BE607585
Length = 443
Score = 195 bits (495), Expect = 5e-50
Identities = 101/145 (69%), Positives = 116/145 (79%), Gaps = 11/145 (7%)
Frame = +1
Query: 372 TLQPIPENPSSTLPS-----SSTTLVAQPRNNN-PISSYSLISSG-ERDDIGRRWPKDEV 424
TLQ IP S++ P+ S+ +L Q NNN P+ + S+++ G E+DD+GRRWPKDEV
Sbjct: 13 TLQVIPSTSSTSTPALPQNPSTYSLNIQNNNNNIPVETNSVLNKGNEKDDVGRRWPKDEV 192
Query: 425 LALINLRC----NNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINKYF 480
LALINLRC NNNNEEKEGN NK PLWERISQGMLELGYKRSAKRCKEKWENINKYF
Sbjct: 193 LALINLRCTSVNNNNNEEKEGN--NKVPLWERISQGMLELGYKRSAKRCKEKWENINKYF 366
Query: 481 RKTKDANRKRSLDSRTCPYFHLLTN 505
RKTKD N+KRSLDSRTCPYFH L++
Sbjct: 367 RKTKDVNKKRSLDSRTCPYFHQLSS 441
Score = 49.3 bits (116), Expect = 5e-06
Identities = 25/64 (39%), Positives = 38/64 (59%), Gaps = 9/64 (14%)
Frame = +1
Query: 104 WTNDEVLALLKIRSSM---------ESWFPDFTWEHVSRKLAEVGYKRSAEKCKEKFEEE 154
W DEVLAL+ +R + E WE +S+ + E+GYKRSA++CKEK+E
Sbjct: 175 WPKDEVLALINLRCTSVNNNNNEEKEGNNKVPLWERISQGMLELGYKRSAKRCKEKWENI 354
Query: 155 SRFF 158
+++F
Sbjct: 355 NKYF 366
>BU549108
Length = 615
Score = 189 bits (481), Expect = 2e-48
Identities = 102/156 (65%), Positives = 115/156 (73%), Gaps = 10/156 (6%)
Frame = -1
Query: 408 SSGERDDIGRRWPKDEVLALINLRC---------NNNNEEKEGNSNNKAPLWERISQGML 458
S +DDIGRRWP+DEVLALI RC NNNNEEKEGN NK PLWERISQGM
Sbjct: 615 SENSKDDIGRRWPRDEVLALIXXRCTXXSSSNNNNNNNEEKEGN--NKGPLWERISQGMS 442
Query: 459 ELGYKRSAKRCKEKWENINKYFRKTKD-ANRKRSLDSRTCPYFHLLTNLYNQGKLVLQSD 517
ELGYKRSAKRCKEKWENINKYFRKTKD N+KRSL+SRTCPYFH L+ LY QGK+V QSD
Sbjct: 441 ELGYKRSAKRCKEKWENINKYFRKTKDNVNKKRSLNSRTCPYFHQLSCLYGQGKIVPQSD 262
Query: 518 QKQESNNVNVPEENVVQEKAKQDENQDGAGESSQVG 553
+++ N +N P N ++ D +Q A ES QVG
Sbjct: 261 EREGKNYLN-PTAN-SGDQVPPDHDQVQADESYQVG 160
Score = 51.6 bits (122), Expect = 9e-07
Identities = 27/77 (35%), Positives = 41/77 (53%), Gaps = 14/77 (18%)
Frame = -1
Query: 104 WTNDEVLALLKIRSSMESWFPDFT--------------WEHVSRKLAEVGYKRSAEKCKE 149
W DEVLAL+ R + S + WE +S+ ++E+GYKRSA++CKE
Sbjct: 582 WPRDEVLALIXXRCTXXSSSNNNNNNNEEKEGNNKGPLWERISQGMSELGYKRSAKRCKE 403
Query: 150 KFEEESRFFNNINHNQN 166
K+E +++F N N
Sbjct: 402 KWENINKYFRKTKDNVN 352
>TC208893 similar to UP|O48592 (O48592) GT2 protein, partial (11%)
Length = 853
Score = 187 bits (475), Expect = 1e-47
Identities = 109/185 (58%), Positives = 128/185 (68%), Gaps = 10/185 (5%)
Frame = +3
Query: 379 NPSSTLPSSSTTLVAQPRNNNPISSYSLISS-------GERDDIGRRWPKDEVLALINLR 431
NPSS+L S + + P +N +S+Y S+ +DDIGRRWP+DEVLALINLR
Sbjct: 9 NPSSSLNSHNNII---PLESNSVSTYKPTSTTPMASSENSKDDIGRRWPRDEVLALINLR 179
Query: 432 CNN--NNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINKYFRKTKD-ANR 488
C + +NEEKEGN K PLWERISQGM LGYKRSAKRCKEKWENINKYFRKTKD N+
Sbjct: 180 CTSLSSNEEKEGN---KGPLWERISQGMSALGYKRSAKRCKEKWENINKYFRKTKDNVNK 350
Query: 489 KRSLDSRTCPYFHLLTNLYNQGKLVLQSDQKQESNNVNVPEENVVQEKAKQDENQDGAGE 548
KRSL+SRTCPYFH L+ LY QGK+V QS+ +E N P N Q D++Q E
Sbjct: 351 KRSLNSRTCPYFHQLSCLYGQGKIVPQSE--REGNYCLNPTPNSGQ-VPPDDDHQVQDDE 521
Query: 549 SSQVG 553
SSQVG
Sbjct: 522 SSQVG 536
Score = 54.3 bits (129), Expect = 1e-07
Identities = 36/113 (31%), Positives = 57/113 (49%), Gaps = 8/113 (7%)
Frame = +3
Query: 62 NLLHPLHPHKDDEDKEQNSTP--SMNNFQIDRDQRQILPQLIDPWTNDEVLALLKIRSSM 119
N + PL + K ++TP S N + D +R W DEVLAL+ +R +
Sbjct: 36 NNIIPLESNSVSTYKPTSTTPMASSENSKDDIGRR---------WPRDEVLALINLRCTS 188
Query: 120 ESWFPDFT------WEHVSRKLAEVGYKRSAEKCKEKFEEESRFFNNINHNQN 166
S + WE +S+ ++ +GYKRSA++CKEK+E +++F N N
Sbjct: 189 LSSNEEKEGNKGPLWERISQGMSALGYKRSAKRCKEKWENINKYFRKTKDNVN 347
>AW458314
Length = 409
Score = 183 bits (464), Expect = 2e-46
Identities = 97/139 (69%), Positives = 110/139 (78%)
Frame = +2
Query: 38 PPFDPYNQQNHPSQHHQLPLQVQPNLLHPLHPHKDDEDKEQNSTPSMNNFQIDRDQRQIL 97
P FDPYN HP HQLPLQ NLLHPLH HKD EDKE+N+T MN +I RDQRQ L
Sbjct: 2 PYFDPYN---HP---HQLPLQPN-NLLHPLH-HKD-EDKEENTTVPMN-LEIQRDQRQQL 151
Query: 98 PQLIDPWTNDEVLALLKIRSSMESWFPDFTWEHVSRKLAEVGYKRSAEKCKEKFEEESRF 157
P+LIDPW NDEVLALL+IRSSMESWFP+ TWEHVSRKL+E+GYK SA KCKEKFEEESR+
Sbjct: 152 PELIDPWNNDEVLALLRIRSSMESWFPELTWEHVSRKLSELGYKMSA*KCKEKFEEESRY 331
Query: 158 FNNINHNQNSFGKNFRFVT 176
FNNIN+ +N+ N +T
Sbjct: 332 FNNINYGENNNNNNNSILT 388
Score = 49.7 bits (117), Expect = 3e-06
Identities = 26/62 (41%), Positives = 37/62 (58%)
Frame = +2
Query: 419 WPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINK 478
W DEVLAL+ +R + + E WE +S+ + ELGYK SA +CKEK+E ++
Sbjct: 170 WNNDEVLALLRIRSSMESWFPELT-------WEHVSRKLSELGYKMSA*KCKEKFEEESR 328
Query: 479 YF 480
YF
Sbjct: 329 YF 334
>TC207639 similar to UP|O48592 (O48592) GT2 protein, partial (24%)
Length = 1689
Score = 155 bits (393), Expect = 3e-38
Identities = 108/338 (31%), Positives = 165/338 (47%), Gaps = 57/338 (16%)
Frame = +1
Query: 269 MHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKELELMAHEQAIAGDRQAHIIQFLNKFS 328
+ + +E + R+ E+ +++EAW+ QEM ++N+E EL+ E++ A + A +I FL + S
Sbjct: 31 VQKRFLEAIDNREREQVAQQEAWRIQEMARINREHELLVQERSTAAAKNAAVIAFLQQLS 210
Query: 329 TSANSSSLTSMSTQL-----------------QAYLATLTSNSSSSTLHSQNPNPETLKK 371
+S+ + Q Q + SNS + + N +
Sbjct: 211 GQHQNSTTQANFPQQPPPQKVTIVQPPPQQAPQPQAPLVLSNSDNMEIQKMNNGHSVVTA 390
Query: 372 TLQPIPE-NPSSTLPSSSTTLVAQPRNNNPISSYSLISSGERDDIGRRWPKDEVLALINL 430
T+ P++T +++T + P SS S +SS RWPK EV ALI L
Sbjct: 391 TVTATATPTPTTTFVAAATAITTTP------SSLSSLSSS-------RWPKTEVHALIRL 531
Query: 431 RCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINKYFRKTKDANRKR 490
R + E K + K PLWE IS GML LGY RSAKRCKEKWENINKYF+K K++N++R
Sbjct: 532 R--TSLETKYQENGPKTPLWEDISAGMLRLGYNRSAKRCKEKWENINKYFKKVKESNKQR 705
Query: 491 SLDSRTCPYFHLLTNLYNQGK---------------------LVLQSDQK---------- 519
DS+TCPYF+ L LY + L++Q +Q+
Sbjct: 706 REDSKTCPYFNELEALYKEKSKTQNPFGASFHNMKPHEMMEPLMVQPEQQWRPPSQYEQG 885
Query: 520 --QESNN------VNVPEENVVQEKAKQDENQDGAGES 549
+E+NN E+ +E+ ++DEN++G ES
Sbjct: 886 AAKENNNNERKEREEEEEDEEEEEEEEEDENEEGDLES 999
Score = 62.0 bits (149), Expect = 7e-10
Identities = 48/189 (25%), Positives = 87/189 (45%), Gaps = 16/189 (8%)
Frame = +1
Query: 104 WTNDEVLALLKIRSSMESWFPDF-----TWEHVSRKLAEVGYKRSAEKCKEKFEEESRFF 158
W EV AL+++R+S+E+ + + WE +S + +GY RSA++CKEK+E +++F
Sbjct: 496 WPKTEVHALIRLRTSLETKYQENGPKTPLWEDISAGMLRLGYNRSAKRCKEKWENINKYF 675
Query: 159 NNI---NHNQNSFGKNFRFVTELEEVYQGGGGENNKNLVEAEKQNEVQDKMDPHEEDSRM 215
+ N + K + ELE +Y+ +N A N M PHE M
Sbjct: 676 KKVKESNKQRREDSKTCPYFNELEALYK--EKSKTQNPFGASFHN-----MKPHE---MM 825
Query: 216 DDVLVSKKS--------EEEVVEKGTTNDEKKRKRSGDDRFEVFKGFCESVVKKMMDQQE 267
+ ++V + E+ ++ N+ K+R+ +D E ++ D+ E
Sbjct: 826 EPLMVQPEQQWRPPSQYEQGAAKENNNNERKEREEEEEDE--------EEEEEEEEDENE 981
Query: 268 EMHNKLIED 276
E + +ED
Sbjct: 982 EGDLESVED 1008
>TC220820 similar to UP|O48592 (O48592) GT2 protein, partial (11%)
Length = 646
Score = 155 bits (392), Expect = 4e-38
Identities = 82/121 (67%), Positives = 90/121 (73%)
Frame = +3
Query: 436 NEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINKYFRKTKDANRKRSLDSR 495
NEEKEGN NK PLWERISQGM EL YKRSAKRCKEKWENINKYFRKTKD +KRSLDSR
Sbjct: 12 NEEKEGN--NKVPLWERISQGMSELRYKRSAKRCKEKWENINKYFRKTKDITKKRSLDSR 185
Query: 496 TCPYFHLLTNLYNQGKLVLQSDQKQESNNVNVPEENVVQEKAKQDENQDGAGESSQVGPP 555
TCPYFH L++LYNQGKLVLQS+ N P++N E+ K D+ SSQVG
Sbjct: 186 TCPYFHQLSSLYNQGKLVLQSESHL---NNTPPDQN--PEQVKPDQTTQAHESSSQVGSG 350
Query: 556 S 556
S
Sbjct: 351 S 353
Score = 43.1 bits (100), Expect = 3e-04
Identities = 29/99 (29%), Positives = 52/99 (52%), Gaps = 11/99 (11%)
Frame = +3
Query: 128 WEHVSRKLAEVGYKRSAEKCKEKFEEESRFF---NNINHNQNSFGKNFRFVTELEEVYQG 184
WE +S+ ++E+ YKRSA++CKEK+E +++F +I ++ + + +L +Y
Sbjct: 48 WERISQGMSELRYKRSAKRCKEKWENINKYFRKTKDITKKRSLDSRTCPYFHQLSSLYNQ 227
Query: 185 G----GGENNKNLVEAEKQNEVQDKMD----PHEEDSRM 215
G E++ N + QN Q K D HE S++
Sbjct: 228 GKLVLQSESHLNNTPPD-QNPEQVKPDQTTQAHESSSQV 341
>TC210182 similar to UP|Q9AVE4 (Q9AVE4) DNA-binding protein DF1, partial
(13%)
Length = 918
Score = 120 bits (301), Expect = 2e-27
Identities = 66/160 (41%), Positives = 91/160 (56%), Gaps = 26/160 (16%)
Frame = +2
Query: 418 RWPKDEVLALINLRC--------NNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRC 469
RWPKDEV ALI LR NNNN +G+ K PLWE IS M LGY RSAKRC
Sbjct: 104 RWPKDEVEALIRLRTQIDVQAQWNNNNNNNDGS---KGPLWEEISSAMKSLGYDRSAKRC 274
Query: 470 KEKWENINKYFRKTKDANRKRSLDSRTCPYFHLLTNLYNQGK------------------ 511
KEKWENINKYF++ K+ ++++ DS+TCPY+H L LY++
Sbjct: 275 KEKWENINKYFKRIKEKSKRKPQDSKTCPYYHHLEALYSKKPKKVDLGHELKPEELLMHI 454
Query: 512 LVLQSDQKQESNNVNVPEENVVQEKAKQDENQDGAGESSQ 551
+V QS ++Q+ + ++ E A++D+NQ + S+
Sbjct: 455 MVSQSQEQQQQQEMQTQTQS-PSEDAERDQNQGDNEDQSE 571
Score = 47.4 bits (111), Expect = 2e-05
Identities = 37/164 (22%), Positives = 73/164 (43%), Gaps = 21/164 (12%)
Frame = +2
Query: 104 WTNDEVLALLKIRSSME---SWFPDFT---------WEHVSRKLAEVGYKRSAEKCKEKF 151
W DEV AL+++R+ ++ W + WE +S + +GY RSA++CKEK+
Sbjct: 107 WPKDEVEALIRLRTQIDVQAQWNNNNNNNDGSKGPLWEEISSAMKSLGYDRSAKRCKEKW 286
Query: 152 EEESRFFNNINHNQN---SFGKNFRFVTELEEVYQGGGGENNKNLVEAEKQNEVQDKMDP 208
E +++F I K + LE +Y + K+ ++ ++ P
Sbjct: 287 ENINKYFKRIKEKSKRKPQDSKTCPYYHHLEALYS-----------KKPKKVDLGHELKP 433
Query: 209 HEEDSRMDDVLVSKKSEEEVVEKGTT------NDEKKRKRSGDD 246
E + ++VS+ E++ ++ T D ++ + GD+
Sbjct: 434 EE---LLMHIMVSQSQEQQQQQEMQTQTQSPSEDAERDQNQGDN 556
>BE610885
Length = 247
Score = 119 bits (299), Expect = 3e-27
Identities = 55/81 (67%), Positives = 67/81 (81%), Gaps = 4/81 (4%)
Frame = +2
Query: 103 PWTNDEVLALLKIRSSMESWFPDFTWEHVSRKLAEVGYKRSAEKCKEKFEEESRFFNNIN 162
PW NDEVLALL+IRSSMESWFP+ TWEHVSRKLAE+GYKRSAEKCKEKFE+ESR+ NNI
Sbjct: 2 PWNNDEVLALLRIRSSMESWFPELTWEHVSRKLAELGYKRSAEKCKEKFEDESRYRNNIK 181
Query: 163 HNQNSF----GKNFRFVTELE 179
+ +N + +R ++EL+
Sbjct: 182 YRENDYSYHNSDRYRCLSELD 244
Score = 52.8 bits (125), Expect = 4e-07
Identities = 27/66 (40%), Positives = 40/66 (59%)
Frame = +2
Query: 419 WPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINK 478
W DEVLAL+ +R + + E WE +S+ + ELGYKRSA++CKEK+E+ ++
Sbjct: 5 WNNDEVLALLRIRSSMESWFPELT-------WEHVSRKLAELGYKRSAEKCKEKFEDESR 163
Query: 479 YFRKTK 484
Y K
Sbjct: 164 YRNNIK 181
>CA937765
Length = 391
Score = 117 bits (293), Expect = 1e-26
Identities = 70/104 (67%), Positives = 77/104 (73%), Gaps = 4/104 (3%)
Frame = +1
Query: 1 MYDGVPDQFHQFITPRTSSSSLPLHLPFPL---STPNNTFPP-FDPYNQQNHPSQHHQLP 56
M+DGVPDQFHQFITPRTS PLHLPFPL TPN TFP FDPY N+PS HQLP
Sbjct: 112 MFDGVPDQFHQFITPRTSQ---PLHLPFPLHASGTPNTTFPSNFDPY---NNPS--HQLP 267
Query: 57 LQVQPNLLHPLHPHKDDEDKEQNSTPSMNNFQIDRDQRQILPQL 100
LQ NLLHPLH HK DE+KE+N+T NF+I RDQRQ LP+L
Sbjct: 268 LQPN-NLLHPLH-HK-DEEKEENTTTVPMNFEIQRDQRQQLPEL 390
>BQ253306
Length = 422
Score = 107 bits (268), Expect = 1e-23
Identities = 58/140 (41%), Positives = 84/140 (59%), Gaps = 20/140 (14%)
Frame = +1
Query: 426 ALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINKYFRKTKD 485
ALI LR + +EK + K PLWE IS M +LGY R+AKRCKEKWENINKYF+K K+
Sbjct: 4 ALIKLR--TSMDEKYQENGPKGPLWEEISASMKKLGYNRNAKRCKEKWENINKYFKKVKE 177
Query: 486 ANRKRSLDSRTCPYFHLLTNLYNQ-----------------GKLVLQSDQK---QESNNV 525
+N++R DS+TCPYFH L LY Q L++Q +Q+ Q+ ++
Sbjct: 178 SNKRRPEDSKTCPYFHQLDALYRQKHKAEESTAAAKAESAVAPLMVQPEQQWPPQQQDDR 357
Query: 526 NVPEENVVQEKAKQDENQDG 545
++ E++ E + +E ++G
Sbjct: 358 DITMEDMENEDDEYEEEREG 417
Score = 50.8 bits (120), Expect = 2e-06
Identities = 36/149 (24%), Positives = 72/149 (48%), Gaps = 14/149 (9%)
Frame = +1
Query: 111 ALLKIRSSMESWFPDF-----TWEHVSRKLAEVGYKRSAEKCKEKFEEESRFFNNI---N 162
AL+K+R+SM+ + + WE +S + ++GY R+A++CKEK+E +++F + N
Sbjct: 4 ALIKLRTSMDEKYQENGPKGPLWEEISASMKKLGYNRNAKRCKEKWENINKYFKKVKESN 183
Query: 163 HNQNSFGKNFRFVTELEEVY-QGGGGENNKNLVEAEK-----QNEVQDKMDPHEEDSRMD 216
+ K + +L+ +Y Q E + +AE + + + P ++D R
Sbjct: 184 KRRPEDSKTCPYFHQLDALYRQKHKAEESTAAAKAESAVAPLMVQPEQQWPPQQQDDR-- 357
Query: 217 DVLVSKKSEEEVVEKGTTNDEKKRKRSGD 245
D+ + E+ DE + +R G+
Sbjct: 358 DITMEDMENED--------DEYEEEREGE 420
>BQ296282 homologue to GP|18182309|gb GT-2 factor {Glycine max}, partial
(33%)
Length = 413
Score = 99.0 bits (245), Expect = 5e-21
Identities = 44/71 (61%), Positives = 53/71 (73%)
Frame = +3
Query: 439 KEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINKYFRKTKDANRKRSLDSRTCP 498
K S K PLWE IS M ++GY R+AKRCKEKWENINKYF+K K++N+KR DS+TCP
Sbjct: 6 KYQESGPKGPLWEEISALMRKMGYNRNAKRCKEKWENINKYFKKVKESNKKRPEDSKTCP 185
Query: 499 YFHLLTNLYNQ 509
YFH L LY +
Sbjct: 186 YFHHLEALYRE 218
Score = 43.5 bits (101), Expect = 2e-04
Identities = 31/117 (26%), Positives = 54/117 (45%), Gaps = 3/117 (2%)
Frame = +3
Query: 128 WEHVSRKLAEVGYKRSAEKCKEKFEEESRFFNNI---NHNQNSFGKNFRFVTELEEVYQG 184
WE +S + ++GY R+A++CKEK+E +++F + N + K + LE +Y+
Sbjct: 39 WEEISALMRKMGYNRNAKRCKEKWENINKYFKKVKESNKKRPEDSKTCPYFHHLEALYR- 215
Query: 185 GGGENNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRK 241
E NK + + ++ + M E+ LV EV T ND R+
Sbjct: 216 ---EKNKGEGQMKPESMMAPLMVQPEQQWPPQTTLV---VPPEVTMGDTQNDPMDRQ 368
>TC210062 GT-2 factor [Glycine max]
Length = 712
Score = 87.8 bits (216), Expect = 1e-17
Identities = 50/167 (29%), Positives = 92/167 (54%)
Frame = +3
Query: 217 DVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIED 276
D+L + S E+ T + + RKR +K F E ++K+++++QEE+ + +E
Sbjct: 252 DLLSNSSSSSTSSEETPTTEARGRKRK-----RKWKDFFERLMKEVIEKQEELQRRFLEA 416
Query: 277 MVKRDEEKFSREEAWKKQEMEKMNKELELMAHEQAIAGDRQAHIIQFLNKFSTSANSSSL 336
+ KR++E+ REEAW+ QEM+++N+E E++A E++IA + A ++ FL K + ++
Sbjct: 417 IEKREQERVVREEAWRMQEMQRINREREILAQERSIAAAKDAAVMTFLQKIAEHQQQETI 596
Query: 337 TSMSTQLQAYLATLTSNSSSSTLHSQNPNPETLKKTLQPIPENPSST 383
L +N++S T+ Q P P+T T P P+ +T
Sbjct: 597 N--------LEPALNNNNNSITVVPQQPVPQT-TTTPTPTPQQAQTT 710
>AW706958 homologue to GP|3056598|gb|A T1F9.19 {Arabidopsis thaliana},
partial (2%)
Length = 424
Score = 86.7 bits (213), Expect = 3e-17
Identities = 53/103 (51%), Positives = 61/103 (58%), Gaps = 9/103 (8%)
Frame = +1
Query: 1 MYDGVPDQFHQFITPRTSSSSLPLHLPFPL------STPNNTFPPFDPYNQQNHPSQHHQ 54
M+DG PDQFHQFI PRT +LPLHL FPL + P NTF PFDPYN PS HH
Sbjct: 139 MFDGAPDQFHQFIAPRT---TLPLHLSFPLHHHASSTPPPNTFLPFDPYN----PSSHHL 297
Query: 55 LPLQVQPNLLHP---LHPHKDDEDKEQNSTPSMNNFQIDRDQR 94
LP +LLHP HK ++DK + NN +I RDQR
Sbjct: 298 LPSLQTNHLLHPPTTSPTHKHEQDKVIAPIVN-NNEEIQRDQR 423
>NP426645 GT-2 factor [Glycine max]
Length = 767
Score = 80.5 bits (197), Expect = 2e-15
Identities = 39/102 (38%), Positives = 64/102 (62%)
Frame = +1
Query: 407 ISSGERDDIGRRWPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSA 466
+ G++ G RWP+ E LAL+ +R + + ++ + K PLWE +S+ + ELGY R+A
Sbjct: 97 VEEGDKSFGGNRWPRQETLALLKIRSDMDVAFRDASV--KGPLWEEVSRKLAELGYHRNA 270
Query: 467 KRCKEKWENINKYFRKTKDANRKRSLDSRTCPYFHLLTNLYN 508
K+CKEK+EN+ KY ++TK+ +S + +T +F L L N
Sbjct: 271 KKCKEKFENVYKYHKRTKEGRSGKS-EGKTYRFFDQLQALEN 393
Score = 72.8 bits (177), Expect = 4e-13
Identities = 42/97 (43%), Positives = 57/97 (58%), Gaps = 7/97 (7%)
Frame = +1
Query: 104 WTNDEVLALLKIRSSMESWFPDFT-----WEHVSRKLAEVGYKRSAEKCKEKFEEESRFF 158
W E LALLKIRS M+ F D + WE VSRKLAE+GY R+A+KCKEKFE ++
Sbjct: 133 WPRQETLALLKIRSDMDVAFRDASVKGPLWEEVSRKLAELGYHRNAKKCKEKFENVYKYH 312
Query: 159 NNINHNQN--SFGKNFRFVTELEEVYQGGGGENNKNL 193
++ S GK +RF +L+ + ENN ++
Sbjct: 313 KRTKEGRSGKSEGKTYRFFDQLQAL------ENNPSI 405
>AW596787 similar to GP|18182311|gb| GT-2 factor {Glycine max}, partial (34%)
Length = 411
Score = 78.2 bits (191), Expect = 9e-15
Identities = 39/92 (42%), Positives = 61/92 (65%)
Frame = +1
Query: 418 RWPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENIN 477
RWP+ E LAL+ +R + + ++ S+ K PLWE +++ + ELGY RSAK+CKEK+EN+
Sbjct: 1 RWPRQETLALLKIRSDMDAVFRD--SSLKGPLWEEVARKLSELGYHRSAKKCKEKFENVY 174
Query: 478 KYFRKTKDANRKRSLDSRTCPYFHLLTNLYNQ 509
KY ++TK+ +R + +T +F L L NQ
Sbjct: 175 KYHKRTKE-SRSGKHEGKTYKFFDQLQALENQ 267
Score = 69.7 bits (169), Expect = 3e-12
Identities = 36/83 (43%), Positives = 52/83 (62%), Gaps = 7/83 (8%)
Frame = +1
Query: 104 WTNDEVLALLKIRSSMESWFPDFT-----WEHVSRKLAEVGYKRSAEKCKEKFEEESRFF 158
W E LALLKIRS M++ F D + WE V+RKL+E+GY RSA+KCKEKFE ++
Sbjct: 4 WPRQETLALLKIRSDMDAVFRDSSLKGPLWEEVARKLSELGYHRSAKKCKEKFENVYKYH 183
Query: 159 NNINHNQNS--FGKNFRFVTELE 179
+++ GK ++F +L+
Sbjct: 184 KRTKESRSGKHEGKTYKFFDQLQ 252
>TC222084 similar to UP|O48592 (O48592) GT2 protein, partial (13%)
Length = 565
Score = 65.1 bits (157), Expect = 8e-11
Identities = 35/93 (37%), Positives = 58/93 (61%), Gaps = 1/93 (1%)
Frame = +1
Query: 442 NSNNKAPL-WERISQGMLELGYKRSAKRCKEKWENINKYFRKTKDANRKRSLDSRTCPYF 500
N+N ++PL +++S+ + ELGY RSAK+CKEK+ENI KY R+TK+ +S ++T +F
Sbjct: 178 NANPQSPLSGKQVSRKLSELGYNRSAKKCKEKFENIYKYHRRTKEGRFGKSNGAKTYRFF 357
Query: 501 HLLTNLYNQGKLVLQSDQKQESNNVNVPEENVV 533
L + L+ + +NN NV ++ V+
Sbjct: 358 EQLEAIDGNHSLLPPTTTDNNNNNNNVGDDVVL 456
Score = 47.4 bits (111), Expect(2) = 5e-07
Identities = 23/56 (41%), Positives = 35/56 (62%), Gaps = 3/56 (5%)
Frame = +1
Query: 129 EHVSRKLAEVGYKRSAEKCKEKFEEESRFFNNINH---NQNSFGKNFRFVTELEEV 181
+ VSRKL+E+GY RSA+KCKEKFE ++ +++ K +RF +LE +
Sbjct: 208 KQVSRKLSELGYNRSAKKCKEKFENIYKYHRRTKEGRFGKSNGAKTYRFFEQLEAI 375
Score = 35.8 bits (81), Expect = 0.052
Identities = 28/90 (31%), Positives = 41/90 (45%), Gaps = 7/90 (7%)
Frame = +2
Query: 386 SSSTTLVAQPRNNNPISSYSLISSGERDD---IGRRWPKDEVLALINLRCNNNNEEKEGN 442
S T L + +S S GE DD RWP++E +AL+N+R + + +
Sbjct: 5 SQETPLENADGGSAAVSDGSKAEHGEDDDRNPAANRWPREETMALLNIR-SEMDVAFQRT 181
Query: 443 SNNKAPLWERISQGMLE----LGYKRSAKR 468
KAP SQG +G +RSA+R
Sbjct: 182 QTLKAPSLGNKSQGNYRS*VIIGVRRSARR 271
Score = 24.6 bits (52), Expect(2) = 5e-07
Identities = 10/20 (50%), Positives = 13/20 (65%)
Frame = +2
Query: 104 WTNDEVLALLKIRSSMESWF 123
W +E +ALL IRS M+ F
Sbjct: 113 WPREETMALLNIRSEMDVAF 172
>BQ741940 homologue to GP|18182311|gb| GT-2 factor {Glycine max}, partial
(25%)
Length = 431
Score = 62.4 bits (150), Expect = 5e-10
Identities = 33/93 (35%), Positives = 51/93 (54%), Gaps = 10/93 (10%)
Frame = +1
Query: 448 PLWERISQGMLELGYKRSAKRCKEKWENINKYFRKTKDANRKRSLDSRTCPYFHLLTNLY 507
PLWE +++ + ELGY RSAK+CKEK+EN+ KY ++TK+ R + +T +F L L
Sbjct: 1 PLWEEVARKLSELGYHRSAKKCKEKFENVYKYHKRTKE-GRSGKHEGKTYKFFDQLQALE 177
Query: 508 NQGKLVL----------QSDQKQESNNVNVPEE 530
NQ + S+ + NN+ P +
Sbjct: 178 NQFTVSSYFSKTTTHFGNSNNNNKHNNITTPNQ 276
Score = 51.2 bits (121), Expect = 1e-06
Identities = 23/54 (42%), Positives = 35/54 (64%), Gaps = 2/54 (3%)
Frame = +1
Query: 128 WEHVSRKLAEVGYKRSAEKCKEKFEEESRFFNNINHNQNS--FGKNFRFVTELE 179
WE V+RKL+E+GY RSA+KCKEKFE ++ ++ GK ++F +L+
Sbjct: 7 WEEVARKLSELGYHRSAKKCKEKFENVYKYHKRTKEGRSGKHEGKTYKFFDQLQ 168
>BU549183 similar to GP|18182311|gb GT-2 factor {Glycine max}, partial (58%)
Length = 512
Score = 48.9 bits (115), Expect = 6e-06
Identities = 23/52 (44%), Positives = 35/52 (67%)
Frame = +1
Query: 457 MLELGYKRSAKRCKEKWENINKYFRKTKDANRKRSLDSRTCPYFHLLTNLYN 508
+ ELGY R+AK+CKEK+EN+ KY ++TK+ +S + +T +F L L N
Sbjct: 1 LAELGYNRNAKKCKEKFENVYKYHKRTKEGRSGKS-EGKTYRFFDQLQALEN 153
Score = 42.4 bits (98), Expect = 6e-04
Identities = 20/47 (42%), Positives = 30/47 (63%), Gaps = 2/47 (4%)
Frame = +1
Query: 135 LAEVGYKRSAEKCKEKFEEESRFFNNINHNQN--SFGKNFRFVTELE 179
LAE+GY R+A+KCKEKFE ++ ++ S GK +RF +L+
Sbjct: 1 LAELGYNRNAKKCKEKFENVYKYHKRTKEGRSGKSEGKTYRFFDQLQ 141
>TC229985
Length = 777
Score = 43.5 bits (101), Expect = 2e-04
Identities = 25/93 (26%), Positives = 44/93 (46%)
Frame = +2
Query: 419 WPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINK 478
W +EV LI++R N+ + + LWE ISQ +L G RS +CK W ++ +
Sbjct: 110 WKHEEVKKLIDMRGELNDRFQV--VKGRMALWEEISQNLLANGISRSPGQCKSLWTSLLQ 283
Query: 479 YFRKTKDANRKRSLDSRTCPYFHLLTNLYNQGK 511
+ + K+ + + + PYF + + K
Sbjct: 284 KYEEVKNEKKNK----KKWPYFEDMERILADNK 370
Score = 31.2 bits (69), Expect = 1.3
Identities = 22/101 (21%), Positives = 42/101 (40%), Gaps = 5/101 (4%)
Frame = +2
Query: 86 NFQIDRDQRQILPQLIDPWTNDEVLALLKIRSSMESWFPDFT-----WEHVSRKLAEVGY 140
N + QR+I W ++EV L+ +R + F WE +S+ L G
Sbjct: 74 NPNLQNQQREIT------WKHEEVKKLIDMRGELNDRFQVVKGRMALWEEISQNLLANGI 235
Query: 141 KRSAEKCKEKFEEESRFFNNINHNQNSFGKNFRFVTELEEV 181
RS +CK + + + + N+ K + + ++E +
Sbjct: 236 SRSPGQCKSLWTSLLQKYEEVK-NEKKNKKKWPYFEDMERI 355
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.309 0.127 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,715,101
Number of Sequences: 63676
Number of extensions: 392378
Number of successful extensions: 4005
Number of sequences better than 10.0: 166
Number of HSP's better than 10.0 without gapping: 3584
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3818
length of query: 557
length of database: 12,639,632
effective HSP length: 102
effective length of query: 455
effective length of database: 6,144,680
effective search space: 2795829400
effective search space used: 2795829400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC135565.15


