

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135505.8 + phase: 0
(454 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
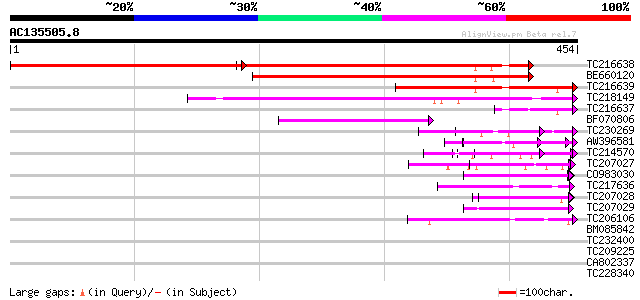
Score E
Sequences producing significant alignments: (bits) Value
TC216638 similar to PIR|D96602|D96602 nucleolar protein [importe... 395 0.0
BE660120 similar to GP|3860319|emb nucleolar protein {Cicer arie... 362 e-100
TC216639 similar to UP|Q9ZRW0 (Q9ZRW0) Nucleolar protein (Fragme... 205 3e-53
TC218149 similar to UP|Q70I27 (Q70I27) SAR DNA-binding protein-l... 194 9e-50
TC216637 homologue to UP|Q9ZRW0 (Q9ZRW0) Nucleolar protein (Frag... 74 2e-13
BF070806 70 2e-12
TC230269 weakly similar to UP|WSC2_YEAST (P53832) Cell wall inte... 63 2e-10
AW396581 61 9e-10
TC214570 homologue to PIR|T05766|T05766 peptidylprolyl isomerase... 61 1e-09
TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%) 51 9e-07
CO983030 46 4e-05
TC217636 homologue to UP|O04235 (O04235) Transcription factor, p... 45 7e-05
TC207028 similar to UP|Q9LW95 (Q9LW95) KED, partial (13%) 44 2e-04
TC207029 weakly similar to UP|Q7R8E7 (Q7R8E7) Ran-binding protei... 43 3e-04
TC206106 weakly similar to UP|Q7RK82 (Q7RK82) Maebl (Fragment), ... 42 7e-04
BM085842 similar to GP|11994428|dbj nucleolar protein {Arabidops... 41 0.001
TC232400 weakly similar to UP|Q8W2K5 (Q8W2K5) Phragmoplastin-int... 40 0.002
TC209225 similar to GB|CAC08342.1|10178284|ATF8L15 rev interacti... 40 0.002
CA802337 40 0.002
TC228340 similar to UP|Q9CJT2 (Q9CJT2) OppB, partial (5%) 40 0.002
>TC216638 similar to PIR|D96602|D96602 nucleolar protein [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(84%)
Length = 1791
Score = 395 bits (1016), Expect(2) = 0.0
Identities = 207/244 (84%), Positives = 222/244 (90%), Gaps = 6/244 (2%)
Frame = +2
Query: 182 KASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARL 241
KASMGQDLSPVDLINVH FAQRVMDLS+YR+ L DYL KMNDIAPNL +L+G+ VGARL
Sbjct: 875 KASMGQDLSPVDLINVHQFAQRVMDLSEYRKNLYDYLVAKMNDIAPNLATLIGEVVGARL 1054
Query: 242 ISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRM 301
ISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASA+NKGRM
Sbjct: 1055 ISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASAKNKGRM 1234
Query: 302 ARYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESV 361
ARYLANKCSIASRIDCF+E+GT+ FGEKLREQVEERLDFYDKGVAPRKNIDVM++AIES
Sbjct: 1235 ARYLANKCSIASRIDCFAERGTTVFGEKLREQVEERLDFYDKGVAPRKNIDVMQAAIESA 1414
Query: 362 DNKDTEMETE---EVSAKKTKKKKQK--AADGDDMAVDKAAEITNGDA-EDHKSEKKKKK 415
+NKDTEMETE EVS KKTKKKKQK AA+GDDMAVD TNGDA EDHKSEKKKKK
Sbjct: 1415 ENKDTEMETEAPAEVSGKKTKKKKQKAAAAEGDDMAVD-----TNGDAVEDHKSEKKKKK 1579
Query: 416 KEKR 419
+K+
Sbjct: 1580 SKKK 1591
Score = 329 bits (844), Expect(2) = 0.0
Identities = 165/189 (87%), Positives = 176/189 (92%)
Frame = +1
Query: 1 MTDELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQ 60
+TDELRTVLETNLPKVKEGKK+KFSLGV++ KIGS I E TKIP QSNEFV EL+RGVR
Sbjct: 256 LTDELRTVLETNLPKVKEGKKSKFSLGVSDPKIGSQISEVTKIPCQSNEFVSELLRGVRF 435
Query: 61 HFDKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAM 120
HFD FVGDLK GDLEKAQLGL HSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKD+NSF+M
Sbjct: 436 HFDTFVGDLKSGDLEKAQLGLGHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDINSFSM 615
Query: 121 RVREWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEA 180
RVREWYSWHFPELVKIVNDNYLY KVAKFIEDK+KLAEDKI +LTD+VGDEDKAKEIVEA
Sbjct: 616 RVREWYSWHFPELVKIVNDNYLYAKVAKFIEDKAKLAEDKIPALTDIVGDEDKAKEIVEA 795
Query: 181 AKASMGQDL 189
AKASMG+ L
Sbjct: 796 AKASMGRIL 822
>BE660120 similar to GP|3860319|emb nucleolar protein {Cicer arietinum},
partial (38%)
Length = 720
Score = 362 bits (928), Expect = e-100
Identities = 189/232 (81%), Positives = 205/232 (87%), Gaps = 7/232 (3%)
Frame = +2
Query: 195 INVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKC 254
+NV FAQRVMDLS+YRR+L DYL KMNDIAPNL SL+G+ VGARLISHAGSLTNLAKC
Sbjct: 2 VNVQQFAQRVMDLSEYRRKLYDYLVAKMNDIAPNLASLIGEVVGARLISHAGSLTNLAKC 181
Query: 255 PSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASR 314
PSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASA+NKGR+ARYLANKCSIASR
Sbjct: 182 PSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASAKNKGRIARYLANKCSIASR 361
Query: 315 IDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETE--- 371
IDCFSE+GT+ FGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESV+ + T METE
Sbjct: 362 IDCFSERGTTTFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVETEYTAMETEAPV 541
Query: 372 EVSAKKTKKKKQKAA---DGDDMAVDKAAEITNGDA-EDHKSEKKKKKKEKR 419
EVS KK KKKK KA DGD+MA DK E TNGDA EDHKSEK+++K+ KR
Sbjct: 542 EVSGKKAKKKKHKAXVTDDGDNMAXDKITETTNGDALEDHKSEKEEEKEGKR 697
>TC216639 similar to UP|Q9ZRW0 (Q9ZRW0) Nucleolar protein (Fragment), partial
(22%)
Length = 711
Score = 205 bits (522), Expect = 3e-53
Identities = 117/175 (66%), Positives = 131/175 (74%), Gaps = 30/175 (17%)
Frame = +2
Query: 310 SIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEME 369
SIASRIDCFSEKGT+ FG+KLREQVEERLDFYDKGVAPRKNIDVM++AIES +NKDTEME
Sbjct: 2 SIASRIDCFSEKGTTVFGQKLREQVEERLDFYDKGVAPRKNIDVMQAAIESAENKDTEME 181
Query: 370 TE---EVSAKKTKKKKQKAADG-DDMAVDKAAEITNGDA-EDHKSEKKKKKKEKRKLDQE 424
TE EVS KKTKKKKQKAAD DD+A+D TNGDA EDHKSE+KKKKKEKRKL+QE
Sbjct: 182 TEAPAEVSGKKTKKKKQKAADAEDDVAID-----TNGDAVEDHKSERKKKKKEKRKLEQE 346
Query: 425 VEVEDKVVEDGAN-------------------------ADSSKKKKKKSKKKDAE 454
+EV+D+ +DGAN + SKKKKKKSKKKD E
Sbjct: 347 MEVDDQAEDDGANGVTSEQEGTAKKKNKKDSNGEAFEAGEVSKKKKKKSKKKDIE 511
>TC218149 similar to UP|Q70I27 (Q70I27) SAR DNA-binding protein-like protein,
partial (57%)
Length = 1030
Score = 194 bits (492), Expect = 9e-50
Identities = 126/344 (36%), Positives = 184/344 (52%), Gaps = 32/344 (9%)
Frame = +2
Query: 143 YCKVAKFIEDKSKLAE-DKIESLTDLVGDEDKAKEIVEAAKASMGQDLSPVDLINVHMFA 201
Y + K + D+ A D E L E+ E+ EA+ SMG ++ +DL N+
Sbjct: 2 YARAVKLMGDRVNAASLDXXEILP-----EEVEAELKEASVISMGTEIGELDLANIRELC 166
Query: 202 QRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTLQI 261
+V+ LS+YR +L DYL ++MN IAPNL ++VG+ VGARLI+H GSL NLAK P ST+QI
Sbjct: 167 DQVLSLSEYRAQLYDYLKSRMNTIAPNLTAMVGELVGARLIAHGGSLLNLAKQPGSTVQI 346
Query: 262 LGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFSEK 321
LGAEKALFRALKT+ TPKYGLI+H+S IG+A+ + KG+++R LA K ++A R D +
Sbjct: 347 LGAEKALFRALKTKHATPKYGLIYHASLIGQAAPKFKGKISRSLAAKTALAIRCDALGDS 526
Query: 322 GTSAFGEKLREQVEERL---------------------DFYDKG--------VAPRKNID 352
+ G + R ++E RL + YDK + P K +
Sbjct: 527 QDNTMGLENRAKLEARLRNLEGKELGRFAGSAKGKPKIEAYDKDRKKGAGGLITPAKTYN 706
Query: 353 VMKSAI--ESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSE 410
++ + VD+ E E A K K+KK K D D + + E +
Sbjct: 707 PSADSVIGQMVDSAIDEDAQEPSVADKKKEKKDKKKKKDKKEEDATVQADGEEPEPVVVK 886
Query: 411 KKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
K KKKK K E E +++G + ++ +KKKK+ K+ + E
Sbjct: 887 KDKKKKRK-------ETESAELQNGNDDNAGEKKKKRKKQGEQE 997
>TC216637 homologue to UP|Q9ZRW0 (Q9ZRW0) Nucleolar protein (Fragment),
partial (8%)
Length = 450
Score = 73.6 bits (179), Expect = 2e-13
Identities = 49/93 (52%), Positives = 54/93 (57%), Gaps = 27/93 (29%)
Frame = +3
Query: 389 DDMAVDKAAEITNGDA-EDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGAN---------- 437
DDMAVD TNGDA EDHKSEKKKKK EKRKL+QE+EV+D+ +DGAN
Sbjct: 15 DDMAVD-----TNGDAVEDHKSEKKKKK-EKRKLEQEMEVDDQAEDDGANGVTSEQEGTV 176
Query: 438 ----------------ADSSKKKKKKSKKKDAE 454
SKKKKKKSKKKD E
Sbjct: 177 KKKKNKKDSNGEALEAGGESKKKKKKSKKKDTE 275
>BF070806
Length = 414
Score = 70.1 bits (170), Expect = 2e-12
Identities = 35/124 (28%), Positives = 67/124 (53%)
Frame = +2
Query: 216 DYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTLQILGAEKALFRALKTR 275
D++ ++M IAPNL ++VG +V A+L+ AG L +LAK P+ +Q+LGA+K T
Sbjct: 8 DFVESRMGYIAPNLSAIVGSAVAAKLMGTAGGLASLAKMPACNVQLLGAKKKNLAGFSTA 187
Query: 276 GNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFSEKGTSAFGEKLREQVE 335
+ + G + + + R R LA K ++A+R+D + G ++++
Sbjct: 188 TSQFRVGYLEQTEIFQTTPPSLRMRACRLLAAKSTLAARVDSIRGDPSGKTGRAFKDEIH 367
Query: 336 ERLD 339
++++
Sbjct: 368 KKIE 379
>TC230269 weakly similar to UP|WSC2_YEAST (P53832) Cell wall integrity and
stress response component 2 precursor, partial (6%)
Length = 696
Score = 63.2 bits (152), Expect = 2e-10
Identities = 43/109 (39%), Positives = 60/109 (54%), Gaps = 12/109 (11%)
Frame = +2
Query: 358 IESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAE-----------ITNGDAED 406
+E V K + ET EV +K +KKK+K D ++ V + E I +G E
Sbjct: 74 LEKVKVKKVDEETVEVEVEKKEKKKKKKKDKENSEVASSDEEKTEKKKNKNKIEDGSPEL 253
Query: 407 HKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKK-KKSKKKDAE 454
KSEKKKKKK+ + E+ + V+D +NAD S+KKK KK K KDA+
Sbjct: 254 DKSEKKKKKKKDKAAAAAAEISNGKVDD-SNADKSEKKKNKKKKNKDAK 397
Score = 48.1 bits (113), Expect = 8e-06
Identities = 32/103 (31%), Positives = 56/103 (54%), Gaps = 2/103 (1%)
Frame = +2
Query: 328 EKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAK--KTKKKKQKA 385
E+ E E+ + K ++N +V S E + K + + E+ S + K++KKK+K
Sbjct: 104 EETVEVEVEKKEKKKKKKKDKENSEVASSDEEKTEKKKNKNKIEDGSPELDKSEKKKKKK 283
Query: 386 ADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVE 428
D A AAEI+NG +D ++K +KKK K+K +++ + E
Sbjct: 284 KD---KAAAAAAEISNGKVDDSNADKSEKKKNKKKKNKDAKEE 403
>AW396581
Length = 401
Score = 61.2 bits (147), Expect = 9e-10
Identities = 34/95 (35%), Positives = 57/95 (59%), Gaps = 3/95 (3%)
Frame = -1
Query: 363 NKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNG---DAEDHKSEKKKKKKEKR 419
+KD + + E+V K+ K+KK+K D D AE G D ED++ +++KKKK+K+
Sbjct: 305 DKDDKTDVEKVKGKEDKEKKKKEKKVKDDKTD--AEKVKGKEDDGEDNEEKEEKKKKKKK 132
Query: 420 KLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
+ D +++VE ++ D KK+KKK +KKD +
Sbjct: 131 EKDDKIDVEKVKGKEDDGEDEGKKEKKKKEKKDKD 27
Score = 47.8 bits (112), Expect = 1e-05
Identities = 29/86 (33%), Positives = 44/86 (50%)
Frame = -1
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K E + E+ KK KKKK+K D V+K G + K +K+KK K+ + +
Sbjct: 371 KVKEHDGEDAEEKKEKKKKEKKDKDDKTDVEKV----KGKEDKEKKKKEKKVKDDKTDAE 204
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSK 449
+V+ ++ ED + KKKKKK K
Sbjct: 203 KVKGKEDDGEDNEEKEEKKKKKKKEK 126
Score = 43.1 bits (100), Expect = 3e-04
Identities = 28/78 (35%), Positives = 42/78 (52%)
Frame = -1
Query: 349 KNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHK 408
K + K+ E V K+ + E E +K KKKK++ D D+ K E D ED +
Sbjct: 236 KKVKDDKTDAEKVKGKEDDGEDNEEKEEKKKKKKKEKDDKIDVEKVKGKE---DDGED-E 69
Query: 409 SEKKKKKKEKRKLDQEVE 426
+K+KKKKEK+ D+E +
Sbjct: 68 GKKEKKKKEKKDKDEETD 15
Score = 38.9 bits (89), Expect = 0.005
Identities = 22/55 (40%), Positives = 33/55 (60%)
Frame = -1
Query: 398 EITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKD 452
++ D ED + +K+KKKKEK+ D + +VE KV + +KKKK+ K KD
Sbjct: 371 KVKEHDGEDAEEKKEKKKKEKKDKDDKTDVE-KV----KGKEDKEKKKKEKKVKD 222
Score = 37.4 bits (85), Expect = 0.014
Identities = 27/83 (32%), Positives = 39/83 (46%), Gaps = 25/83 (30%)
Frame = -1
Query: 395 KAAEITNGDAEDHKSEKKKKKKE----------KRKLDQEVEVEDKVVEDGA-------- 436
K E DAE+ K +KKK+KK+ K K D+E + ++K V+D
Sbjct: 371 KVKEHDGEDAEEKKEKKKKEKKDKDDKTDVEKVKGKEDKEKKKKEKKVKDDKTDAEKVKG 192
Query: 437 -------NADSSKKKKKKSKKKD 452
N + +KKKKK K+KD
Sbjct: 191 KEDDGEDNEEKEEKKKKKKKEKD 123
>TC214570 homologue to PIR|T05766|T05766 peptidylprolyl isomerase M4E13.20 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(13%)
Length = 944
Score = 60.8 bits (146), Expect = 1e-09
Identities = 41/109 (37%), Positives = 57/109 (51%), Gaps = 9/109 (8%)
Frame = +2
Query: 355 KSAIESVDNKDTEMETEEVSAKKTKKKKQK---AADGDDMAVDKAAEITNGDAEDH---- 407
K ++ VD + E+E E+ KK KKK ++ AA D+ +K + ED
Sbjct: 434 KVKVKKVDEETVEVEVEKKEKKKKKKKNKENSEAASSDEEKTEKKKKKHKDKVEDSSPEL 613
Query: 408 -KSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKK-KKSKKKDAE 454
KSEKKKKKK+ ++ ED +NAD S+KKK KK K KDA+
Sbjct: 614 DKSEKKKKKKKDKEAAAATAEISNGKEDDSNADKSEKKKHKKKKNKDAD 760
Score = 45.8 bits (107), Expect = 4e-05
Identities = 31/104 (29%), Positives = 54/104 (51%), Gaps = 7/104 (6%)
Frame = +2
Query: 332 EQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEME-------TEEVSAKKTKKKKQK 384
E VE ++ +K +KN + ++A S D + TE + E+ S + K +K+K
Sbjct: 461 ETVEVEVEKKEKKKKKKKNKENSEAA--SSDEEKTEKKKKKHKDKVEDSSPELDKSEKKK 634
Query: 385 AADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVE 428
D A AEI+NG +D ++K +KKK K+K +++ + E
Sbjct: 635 KKKKDKEAAAATAEISNGKEDDSNADKSEKKKHKKKKNKDADEE 766
Score = 43.5 bits (101), Expect = 2e-04
Identities = 35/96 (36%), Positives = 46/96 (47%)
Frame = +2
Query: 359 ESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEK 418
E V K + ET EV +K +KKK+K + + N +A EK +KKK+K
Sbjct: 431 EKVKVKKVDEETVEVEVEKKEKKKKKKKNKE-----------NSEAASSDEEKTEKKKKK 577
Query: 419 RKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
K DKV + D S+KK KK KKKD E
Sbjct: 578 HK--------DKVEDSSPELDKSEKK-KK-KKKDKE 655
Score = 43.1 bits (100), Expect = 3e-04
Identities = 29/93 (31%), Positives = 44/93 (47%), Gaps = 12/93 (12%)
Frame = +2
Query: 373 VSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKK------------EKRK 420
V AKK K + + + + K + + E K EKKKKKK E++
Sbjct: 380 VPAKKVKVDEVEEVEKQEKVKVKKVDEETVEVEVEKKEKKKKKKKNKENSEAASSDEEKT 559
Query: 421 LDQEVEVEDKVVEDGANADSSKKKKKKSKKKDA 453
++ + +DKV + D S+KKKKK K K+A
Sbjct: 560 EKKKKKHKDKVEDSSPELDKSEKKKKKKKDKEA 658
Score = 30.8 bits (68), Expect = 1.3
Identities = 17/45 (37%), Positives = 28/45 (61%), Gaps = 1/45 (2%)
Frame = +2
Query: 411 KKKKKKEKRKLDQEVEVE-DKVVEDGANADSSKKKKKKSKKKDAE 454
KK K E +++++ +V+ KV E+ + KK+KKK KKK+ E
Sbjct: 389 KKVKVDEVEEVEKQEKVKVKKVDEETVEVEVEKKEKKKKKKKNKE 523
>TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%)
Length = 1005
Score = 51.2 bits (121), Expect = 9e-07
Identities = 32/87 (36%), Positives = 48/87 (54%), Gaps = 2/87 (2%)
Frame = +3
Query: 369 ETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVE 428
E ++ KK KKKK+ GD V+K N +D + ++KKKK++K K D + + E
Sbjct: 15 EEKKEKDKKEKKKKENKDKGDKTDVEKVKGKENDGEDDEEKKEKKKKEKKDK-DGKTDAE 191
Query: 429 --DKVVEDGANADSSKKKKKKSKKKDA 453
K +DG + + K+KKKK K DA
Sbjct: 192 KIKKKEDDGKDDEEKKEKKKKYDKIDA 272
Score = 50.8 bits (120), Expect = 1e-06
Identities = 46/150 (30%), Positives = 70/150 (46%), Gaps = 18/150 (12%)
Frame = +3
Query: 320 EKGTSAFGEKLREQVEERLDFYDKGVAPRKN-------IDVMKSAIESVDNKDTEMETEE 372
+KG EK++ + + D +K +K D K + D KD E + E+
Sbjct: 66 DKGDKTDVEKVKGKENDGEDDEEKKEKKKKEKKDKDGKTDAEKIKKKEDDGKDDEEKKEK 245
Query: 373 ------VSAKKTKKKKQKAAD-GDDMAVDKAAEITNGDAEDHKSEK---KKKKKEKRKLD 422
+ A K K K+ D G+ DK E +GD E+ K +K K+KKKEK+ D
Sbjct: 246 KKKYDKIDAXKVKGKEDDGKDEGNKEKKDK--EKGDGDGEEKKEKKDKEKEKKKEKKDKD 419
Query: 423 QEVE-VEDKVVEDGANADSSKKKKKKSKKK 451
+E + +++K D D KKKKK KK+
Sbjct: 420 EETDTLKEKGKNDEGEDDEGNKKKKKDKKE 509
Score = 48.1 bits (113), Expect = 8e-06
Identities = 38/132 (28%), Positives = 60/132 (44%)
Frame = +3
Query: 320 EKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTK 379
EK G+ E+ +E+ D + +K+ D ++ K+ E E +E + KK K
Sbjct: 321 EKKDKEKGDGDGEEKKEKKDKEKEKKKEKKDKDEETDTLKE-KGKNDEGEDDEGNKKKKK 497
Query: 380 KKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANAD 439
KK+K D DK G+ ED K E + + ++ +E E EDK + G
Sbjct: 498 DKKEKEKDHKKEKKDKE----EGEKEDSKVEVSVRDIDIEEIKKEGEKEDKGKDGGKEVK 665
Query: 440 SSKKKKKKSKKK 451
KKK+ K KK+
Sbjct: 666 EKKKKEDKDKKE 701
Score = 43.1 bits (100), Expect = 3e-04
Identities = 40/157 (25%), Positives = 69/157 (43%), Gaps = 24/157 (15%)
Frame = +3
Query: 320 EKGTSAFGEKLREQVEERLDFYDKGVAPRK--NIDVMKSAIESVDNKDT----------- 366
+K EK++++ ++ D +K +K ID K + D KD
Sbjct: 165 DKDGKTDAEKIKKKEDDGKDDEEKKEKKKKYDKIDAXKVKGKEDDGKDEGNKEKKDKEKG 344
Query: 367 EMETEEVSAKKTKKKKQKAADGD-DMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEV 425
+ + EE KK K+K++K D D D E D + KKKKK+K++ +++
Sbjct: 345 DGDGEEKKEKKDKEKEKKKEKKDKDEETDTLKEKGKNDEGEDDEGNKKKKKDKKEKEKDH 524
Query: 426 EVEDKVVEDGANADSS----------KKKKKKSKKKD 452
+ E K E+G DS ++ KK+ +K+D
Sbjct: 525 KKEKKDKEEGEKEDSKVEVSVRDIDIEEIKKEGEKED 635
Score = 41.2 bits (95), Expect = 0.001
Identities = 21/55 (38%), Positives = 33/55 (59%), Gaps = 1/55 (1%)
Frame = +3
Query: 401 NGDAEDHKSEKKKKKKEKRKLDQEVEVED-KVVEDGANADSSKKKKKKSKKKDAE 454
+G+ + K +K+KKKKE + + +VE K E+ D KK+KKK +KKD +
Sbjct: 9 DGEEKKEKDKKEKKKKENKDKGDKTDVEKVKGKENDGEDDEEKKEKKKKEKKDKD 173
>CO983030
Length = 505
Score = 45.8 bits (107), Expect = 4e-05
Identities = 28/88 (31%), Positives = 44/88 (49%)
Frame = +3
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K + + ++ KK KKKK+K K + + K +KKKKKK+K+K +
Sbjct: 33 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 212
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSKKK 451
+ + + K + KKKKKK+KKK
Sbjct: 213 KKKKKKKKKKKKKKKKKKKKKKKKNKKK 296
Score = 45.8 bits (107), Expect = 4e-05
Identities = 28/88 (31%), Positives = 44/88 (49%)
Frame = +1
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K + + ++ KK KKKK+K K + + K +KKKKKK+K+K +
Sbjct: 34 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 213
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSKKK 451
+ + + K + KKKKKK+KKK
Sbjct: 214 KKKKKKKKKKKKKKKKKKKKKKKKTKKK 297
Score = 45.4 bits (106), Expect = 5e-05
Identities = 28/88 (31%), Positives = 43/88 (48%)
Frame = +2
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K + + ++ KK KKKK+K K + + K +KKKKKK+K+K +
Sbjct: 23 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 202
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSKKK 451
+ + + K + KKKKKK KKK
Sbjct: 203 KKKKKKKKKKKKKKKKKKKKKKKKKKKK 286
Score = 45.4 bits (106), Expect = 5e-05
Identities = 28/88 (31%), Positives = 43/88 (48%)
Frame = +2
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K + + ++ KK KKKK+K K + + K +KKKKKK+K+K +
Sbjct: 14 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 193
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSKKK 451
+ + + K + KKKKKK KKK
Sbjct: 194 KKKKKKKKKKKKKKKKKKKKKKKKKKKK 277
Score = 43.9 bits (102), Expect = 2e-04
Identities = 27/88 (30%), Positives = 43/88 (48%)
Frame = +2
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K + + ++ KK KKKK+K K + + K +KKKKKK+K+K +
Sbjct: 38 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 217
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSKKK 451
+ + + K + KKKKK+ KKK
Sbjct: 218 KKKKKKKKKKKKKKKKKKKKKKKQKKKK 301
Score = 42.0 bits (97), Expect = 6e-04
Identities = 26/89 (29%), Positives = 43/89 (48%)
Frame = +2
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K + + ++ KK KKKK+K K + + K +KKKKKK+K+K +
Sbjct: 44 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 223
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSKKKD 452
+ + + K + KKK+KK K K+
Sbjct: 224 KKKKKKKKKKKKKKKKKKKKKQKKKKXKN 310
Score = 41.6 bits (96), Expect = 7e-04
Identities = 26/87 (29%), Positives = 42/87 (47%)
Frame = +2
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K + + ++ KK KKKK+K K + + K +KKKKKK+K+K +
Sbjct: 56 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 235
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSKK 450
+ + + K + KKKK K+KK
Sbjct: 236 KKKKKKKKKKKKKKKKKQKKKKXKNKK 316
Score = 40.8 bits (94), Expect = 0.001
Identities = 25/88 (28%), Positives = 44/88 (49%)
Frame = +2
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K + + ++ KK KKKK+K K + + K +KKKKKK+K+K +
Sbjct: 89 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 268
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSKKK 451
+ + + + + N ++KKKK + KK
Sbjct: 269 KKKKKKQKKKKXKNKKNTKKKKXXNMKK 352
Score = 38.5 bits (88), Expect = 0.006
Identities = 25/89 (28%), Positives = 41/89 (45%)
Frame = +1
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K + + ++ KK KKKK+K K + + K +KKKKKK+K+K +
Sbjct: 82 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 261
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSKKKD 452
+ + + K + K KKKK K +
Sbjct: 262 KKKKKKKKTKKKKXXK*KKHKKKKXXKHE 348
Score = 38.1 bits (87), Expect = 0.008
Identities = 26/76 (34%), Positives = 35/76 (45%)
Frame = +1
Query: 376 KKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDG 435
KK KKKK+K K +KKKKKK+K+K ++ + + K +
Sbjct: 4 KKKKKKKKK-----------------------KKKKKKKKKKKKKKKKKKKKKKKKKKKK 114
Query: 436 ANADSSKKKKKKSKKK 451
KKKKKK KKK
Sbjct: 115 KKKKKKKKKKKKKKKK 162
Score = 38.1 bits (87), Expect = 0.008
Identities = 26/76 (34%), Positives = 35/76 (45%)
Frame = +3
Query: 376 KKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDG 435
KK KKKK+K K +KKKKKK+K+K ++ + + K +
Sbjct: 3 KKKKKKKKK-----------------------KKKKKKKKKKKKKKKKKKKKKKKKKKKK 113
Query: 436 ANADSSKKKKKKSKKK 451
KKKKKK KKK
Sbjct: 114 KKKKKKKKKKKKKKKK 161
Score = 38.1 bits (87), Expect = 0.008
Identities = 25/88 (28%), Positives = 41/88 (46%)
Frame = +2
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K + + ++ KK KKKK+K K + + K +KKKKKK+K+K +
Sbjct: 113 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKQK 292
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSKKK 451
+ + ++K + KKK KKK
Sbjct: 293 KKKXKNKKNTKKKKXXNMKKKIXNIKKK 376
>TC217636 homologue to UP|O04235 (O04235) Transcription factor, partial (45%)
Length = 1101
Score = 45.1 bits (105), Expect = 7e-05
Identities = 31/110 (28%), Positives = 49/110 (44%)
Frame = +2
Query: 343 KGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNG 402
K V + D + +E + N+ E E++E + K + + DD D + +G
Sbjct: 314 KKVLENDDDDAVDPHLERIKNEAGEDESDEEDSDFVADKDDEGSPTDDSGADDSDATDSG 493
Query: 403 DAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKD 452
D + EK KK+ K+ L + K +D D KKKK+ KKKD
Sbjct: 494 D----EKEKPAKKESKKDLPSKASTSKKKSKD----DEDGKKKKQKKKKD 619
>TC207028 similar to UP|Q9LW95 (Q9LW95) KED, partial (13%)
Length = 490
Score = 43.9 bits (102), Expect = 2e-04
Identities = 30/94 (31%), Positives = 46/94 (48%), Gaps = 12/94 (12%)
Frame = +1
Query: 371 EEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDK 430
EE KK K+KK++ D D+ + N + ED + KKKKK +K++ +++ + E K
Sbjct: 151 EEKKEKKEKEKKKEKKDKDEETDTLKEKGKNDEGEDDEGNKKKKK-DKKEKEKDHKKEKK 327
Query: 431 VVEDGANADS------------SKKKKKKSKKKD 452
+E+G DS KK K K KD
Sbjct: 328 DIEEGEKEDSKVEASVRDIDIEEKKVTGKDKTKD 429
Score = 42.0 bits (97), Expect = 6e-04
Identities = 21/76 (27%), Positives = 42/76 (54%)
Frame = +1
Query: 376 KKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDG 435
++ +K+K++ + ++ +G+ + K EK+KKK++K K ++ +++K D
Sbjct: 70 RRRRKRKRRTKMKKQTH*REREKMVSGEEKKEKKEKEKKKEKKDKDEETDTLKEKGKNDE 249
Query: 436 ANADSSKKKKKKSKKK 451
D KKKKK KK+
Sbjct: 250 GEDDEGNKKKKKDKKE 297
>TC207029 weakly similar to UP|Q7R8E7 (Q7R8E7) Ran-binding protein, partial
(19%)
Length = 712
Score = 43.1 bits (100), Expect = 3e-04
Identities = 29/88 (32%), Positives = 42/88 (46%)
Frame = +1
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K+ E E +E + KK KKK +K + D K G+ E K E + + + +
Sbjct: 37 KNDEGEDDEEN-KKNKKKDKKEKEKDHKKEKKEDGKEEGEKEKSKVEVSVRDIDIEETTK 213
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSKKK 451
E E EDK +DG KKK+ K KK+
Sbjct: 214 EGEKEDKEKDDGKEVKEKKKKEDKDKKE 297
>TC206106 weakly similar to UP|Q7RK82 (Q7RK82) Maebl (Fragment), partial (7%)
Length = 1040
Score = 41.6 bits (96), Expect = 7e-04
Identities = 38/143 (26%), Positives = 69/143 (47%), Gaps = 7/143 (4%)
Frame = +2
Query: 319 SEKGTSAFGEKLREQV---EERLDFYDKGVAPRKNIDVMKSAI-ESVDNKDTEMETEEVS 374
+EK S +K+ + V EE D A RK +D +K + E+++N + E
Sbjct: 485 AEKAQSGAEDKISQSVSFKEETNVVGDLPEAQRKALDELKRLVQEALNNHELTAPKPEPE 664
Query: 375 AKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVED 434
KKT+ K + + + K E+T E ++E ++KKE +++ EVE V E+
Sbjct: 665 KKKTELKALEKKEEEVSEEKKEVEVTE---EKKEAEVTEEKKEAEVTEEKKEVE--VAEE 829
Query: 435 GANADSSKKKKK---KSKKKDAE 454
+ +++KK+ +KK+ E
Sbjct: 830 KKEVEGTEEKKEVEVAEEKKETE 898
Score = 38.1 bits (87), Expect = 0.008
Identities = 38/152 (25%), Positives = 74/152 (48%), Gaps = 25/152 (16%)
Frame = +2
Query: 328 EKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAAD 387
++L+ V+E L+ ++ AP+ + K+ +++++ K+ E+ E+ + T++KK+
Sbjct: 593 DELKRLVQEALNNHEL-TAPKPEPEKKKTELKALEKKEEEVSEEKKEVEVTEEKKEAEVT 769
Query: 388 GDDMAVDKAAEITNGDAEDHKSEKK---------------KKKKE----KRKLDQEVEVE 428
+ K AE+T E +E+K ++KKE K K + EV E
Sbjct: 770 EEK----KEAEVTEEKKEVEVAEEKKEVEGTEEKKEVEVAEEKKETEVIKEKKEVEVTEE 937
Query: 429 DK---VVEDGANADSSKKKKK---KSKKKDAE 454
K V E+ A+ ++KK+ +KK+AE
Sbjct: 938 KKEVEVTEEKKEAEVIEEKKEVDATEEKKEAE 1033
>BM085842 similar to GP|11994428|dbj nucleolar protein {Arabidopsis
thaliana}, partial (5%)
Length = 428
Score = 40.8 bits (94), Expect = 0.001
Identities = 21/39 (53%), Positives = 25/39 (63%)
Frame = +3
Query: 15 KVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGE 53
K K+ + K SLGV++ KIG I E KIP QSNEF E
Sbjct: 312 KEKKAGQFKISLGVSDPKIGCQISELPKIPCQSNEFDSE 428
>TC232400 weakly similar to UP|Q8W2K5 (Q8W2K5) Phragmoplastin-interacting
protein PHIP1, partial (8%)
Length = 621
Score = 40.4 bits (93), Expect = 0.002
Identities = 22/58 (37%), Positives = 35/58 (59%)
Frame = +2
Query: 363 NKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRK 420
NK+ + ET E KK+K+K +GDD+A NG ++ K++KKK+K++K K
Sbjct: 197 NKEIQTET-EAEGSPNKKRKRKKVEGDDVA-------ANGVSDTPKNKKKKQKQKKNK 346
>TC209225 similar to GB|CAC08342.1|10178284|ATF8L15 rev interacting protein
mis3-like {Arabidopsis thaliana;} , partial (19%)
Length = 779
Score = 40.4 bits (93), Expect = 0.002
Identities = 28/87 (32%), Positives = 43/87 (49%), Gaps = 7/87 (8%)
Frame = +3
Query: 369 ETEEVSAKKTKKKKQKAADG------DDMAVDKAAEITNGDAEDHKSEKKKKKK-EKRKL 421
E +E A+KT + K+K + VDK+ + N A+ S KKK KK KRK
Sbjct: 171 EKQEKQAEKTAENKRKREEAFIPPKEPANLVDKSEDANNNVADMAISLKKKTKKFGKRKS 350
Query: 422 DQEVEVEDKVVEDGANADSSKKKKKKS 448
++ ++ E +V A K KK++S
Sbjct: 351 EENIDAETYIVGSSEQASGKKPKKQRS 431
>CA802337
Length = 381
Score = 40.0 bits (92), Expect = 0.002
Identities = 27/79 (34%), Positives = 37/79 (46%)
Frame = +2
Query: 373 VSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVV 432
V+ KK KKKK+K K +KKKKKK+K+K ++ + + K
Sbjct: 32 VTVKKKKKKKKKK----------------------KKKKKKKKKKKKKKKKKKKKKKKKK 145
Query: 433 EDGANADSSKKKKKKSKKK 451
+ KKKKKK KKK
Sbjct: 146 KKKKKKKKKKKKKKKKKKK 202
>TC228340 similar to UP|Q9CJT2 (Q9CJT2) OppB, partial (5%)
Length = 466
Score = 40.0 bits (92), Expect = 0.002
Identities = 32/113 (28%), Positives = 54/113 (47%), Gaps = 7/113 (6%)
Frame = +2
Query: 349 KNIDVMKSAIESVDNKDT----EMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDA 404
KN ++ K+ + N +T + +T+ KT+KKK K A+ K AEI +
Sbjct: 122 KNTEISKTQKKKTKNAETPKTEKKKTKNAETPKTEKKKTKNAETPKTEKKKNAEIPKIEK 301
Query: 405 EDHKSEKKKKKKEKRKLDQEVEVED---KVVEDGANADSSKKKKKKSKKKDAE 454
+ KS + K K D+E+ E+ K + + +SK++ + SKK AE
Sbjct: 302 KKTKSAEIPLTDGKTKRDREIAKEENKKKTKKPKLSNGTSKQRDEGSKKGVAE 460
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.130 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,488,419
Number of Sequences: 63676
Number of extensions: 177197
Number of successful extensions: 9542
Number of sequences better than 10.0: 264
Number of HSP's better than 10.0 without gapping: 1722
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3505
length of query: 454
length of database: 12,639,632
effective HSP length: 100
effective length of query: 354
effective length of database: 6,272,032
effective search space: 2220299328
effective search space used: 2220299328
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC135505.8


