

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135505.15 - phase: 0 /pseudo
(403 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
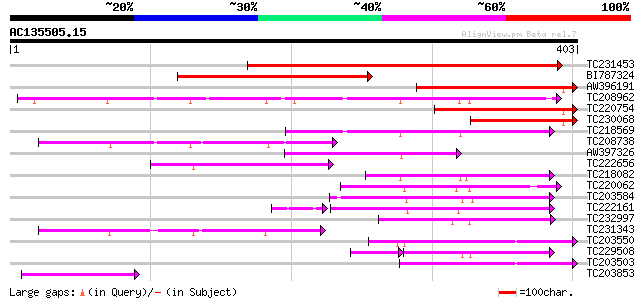
Score E
Sequences producing significant alignments: (bits) Value
TC231453 weakly similar to UP|Q85V22 (Q85V22) Histidine amino ac... 282 2e-76
BI787324 similar to GP|14194151|gb AT5g40780/K1B16_3 {Arabidopsi... 202 3e-52
AW396191 similar to GP|14194151|gb AT5g40780/K1B16_3 {Arabidopsi... 166 2e-41
TC208962 UP|Q9ARG2 (Q9ARG2) Amino acid transporter, complete 152 2e-37
TC220754 similar to UP|Q85V22 (Q85V22) Histidine amino acid tran... 149 2e-36
TC230068 similar to UP|Q85V22 (Q85V22) Histidine amino acid tran... 124 1e-28
TC218569 similar to UP|Q93X14 (Q93X14) Amino acid permease AAP3,... 109 3e-24
TC208738 similar to UP|Q7Y076 (Q7Y076) Amino acid permease 6, pa... 100 2e-21
AW397326 100 2e-21
TC222656 99 3e-21
TC218082 similar to UP|P92934 (P92934) Amino acid permease 6, pa... 89 4e-18
TC220062 weakly similar to UP|Q93X13 (Q93X13) Amino acid permeas... 88 6e-18
TC203584 weakly similar to UP|Q93X15 (Q93X15) Amino acid permeas... 85 5e-17
TC222161 weakly similar to UP|Q9SGD7 (Q9SGD7) T23G18.9, partial ... 77 6e-17
TC232997 weakly similar to UP|Q9ARG2 (Q9ARG2) Amino acid transpo... 82 6e-16
TC231343 similar to UP|Q9FF99 (Q9FF99) Amino acid transporter, p... 75 4e-14
TC203550 weakly similar to UP|Q9SXF7 (Q9SXF7) Amino acid transpo... 72 6e-13
TC229508 similar to UP|Q941E6 (Q941E6) AT5g23810/MRO11_15, parti... 60 6e-13
TC203503 weakly similar to UP|Q9SXF7 (Q9SXF7) Amino acid transpo... 70 1e-12
TC203853 weakly similar to UP|Q9SXF7 (Q9SXF7) Amino acid transpo... 68 6e-12
>TC231453 weakly similar to UP|Q85V22 (Q85V22) Histidine amino acid
transporter, partial (51%)
Length = 811
Score = 282 bits (721), Expect = 2e-76
Identities = 127/224 (56%), Positives = 167/224 (73%)
Frame = +1
Query: 170 LSYSTIAWIASIHRGALPDVQYSSRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTI 229
LSYSTI+W+A + RG + +V Y+ + +T +F IFNALG I+F +AGH V LEIQ+TI
Sbjct: 7 LSYSTISWVACLARGRVENVSYAYKKTTSTDLMFRIFNALGQISFAFAGHAVALEIQATI 186
Query: 230 PSTPEKPSKVSMWRGMIIAYLVVALCYFPVTIFGYRAFGNSVDDNILLSLEKPRWLIIAA 289
PSTPEKPSK+ MW+G I AY++ A+CYFPV + GY AFG V+DN+L+ E+P WLI +A
Sbjct: 187 PSTPEKPSKIPMWKGAIGAYVINAICYFPVALVGYWAFGRDVEDNVLMEFERPAWLIASA 366
Query: 290 NIFVVVHVVGSYQVYAVPVFHMLESFLAEKMNFKPSRFLRFAIRNLYVSITMVLAITFPF 349
N+ V +HVVGSYQVYA+PVF ++ES + ++ F P LR R+ YV+ T+ + +TFPF
Sbjct: 367 NLMVFIHVVGSYQVYAMPVFDLIESMMVKRFKFPPGVALRLVARSAYVAFTLFVGVTFPF 546
Query: 350 FGGLLSFFGGFVFAPTTYFLPCIMWIFIYKPKLFSLSWCANWVS 393
FG LL FFGGF FAPT+YFLP IMW+ I KPK FS +W NW+S
Sbjct: 547 FGDLLGFFGGFGFAPTSYFLPSIMWLIIKKPKRFSTNWFINWIS 678
>BI787324 similar to GP|14194151|gb AT5g40780/K1B16_3 {Arabidopsis thaliana},
partial (31%)
Length = 421
Score = 202 bits (513), Expect = 3e-52
Identities = 88/139 (63%), Positives = 114/139 (81%)
Frame = +3
Query: 120 KKLHEILCDDCEPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIA 179
KK H+ +C +C+ IK T+FI++FA V +VLSHLP FNS+ G+SL AA MSLSYSTIAW+A
Sbjct: 3 KKFHDTVCSNCKNIKLTFFIMIFASVHFVLSHLPDFNSITGVSLAAAVMSLSYSTIAWVA 182
Query: 180 SIHRGALPDVQYSSRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKV 239
S+H+G +VQY + + +G +F FNALG +AF YAGHNV+LEIQ+TIPSTPEKPSKV
Sbjct: 183 SVHKGVQENVQYGYKAKSTSGTVFNFFNALGTVAFAYAGHNVVLEIQATIPSTPEKPSKV 362
Query: 240 SMWRGMIIAYLVVALCYFP 258
MWRG+++AY+VVA+CYFP
Sbjct: 363 PMWRGVVVAYIVVAICYFP 419
>AW396191 similar to GP|14194151|gb AT5g40780/K1B16_3 {Arabidopsis thaliana},
partial (29%)
Length = 444
Score = 166 bits (420), Expect = 2e-41
Identities = 75/125 (60%), Positives = 95/125 (76%), Gaps = 11/125 (8%)
Frame = -3
Query: 290 NIFVVVHVVGSYQVYAVPVFHMLESFLAEKMNFKPSRFLRFAIRNLYVSITMVLAITFPF 349
N+FVV+HV+GSYQ+YA+PVF M+E+ + +++ FKP+ LRF +RN+YV+ TM + ITFPF
Sbjct: 442 NMFVVIHVIGSYQLYAMPVFDMIETVMVKQLRFKPTWQLRFVVRNVYVAFTMFVGITFPF 263
Query: 350 FGGLLSFFGGFVFAPTTYFLPCIMWIFIYKPKLFSLSWCANWV-----------SPIGAL 398
FG LL FFGGF FAPTTYFLPCI+W+ IYKPK FSLSW NW+ SPIG L
Sbjct: 262 FGALLGFFGGFAFAPTTYFLPCIIWLAIYKPKKFSLSWITNWICIIFGLLLMILSPIGGL 83
Query: 399 RQVIL 403
R +IL
Sbjct: 82 RSIIL 68
>TC208962 UP|Q9ARG2 (Q9ARG2) Amino acid transporter, complete
Length = 1789
Score = 152 bits (385), Expect = 2e-37
Identities = 119/443 (26%), Positives = 201/443 (44%), Gaps = 56/443 (12%)
Frame = +1
Query: 6 INIKSRNAKCC-------------YSAFHNVTAMVGAAVLGFPYAMSQLGWGLGITILVL 52
+NI+S +KC + H +TA++G+ VL +A++QLGW G ++ L
Sbjct: 127 MNIQSNYSKCFDDDGRLKRTGTFWMATAHIITAVIGSGVLSLAWAVAQLGWVAGPIVMFL 306
Query: 53 SWICTLYTAWQMIEMH---ESVSGKRFDKYHELSQHAFGERLGLWIVVPQQLMVEVGIDI 109
+ LYT+ + + + +SVSG R Y E G + + Q + + G+ I
Sbjct: 307 FAVVNLYTSNLLTQCYRTGDSVSGHRNYTYMEAVNSILGGKKVKLCGLTQYINL-FGVAI 483
Query: 110 VYMVIGAKSLKKLHEILC-------DDCEPIKTTYFIVLFAFVQYVLSHLPSFNSVAGIS 162
Y + + S+ + C D C Y I F + + S +P F+ V +S
Sbjct: 484 GYTIAASVSMMAIKRSNCYHSSHGKDPCHMSSNGYMIT-FGIAEVIFSQIPDFDQVWWLS 660
Query: 163 LVAAAMSLSYSTIAWIASI--------HRGALPDVQYSSRYSTKAGN------IFGIFNA 208
+VAA MS +YS++ + +G+L + + T+AG I+ A
Sbjct: 661 IVAAIMSFTYSSVGLSLGVAKVAENKSFKGSLMGISIGT--VTQAGTVTSTQKIWRSLQA 834
Query: 209 LGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWRGMIIAYLVVALCYFPVTIFGYRAFG 268
LG +AF Y+ +++EIQ TI S P + +M + ++ V + Y GY AFG
Sbjct: 835 LGAMAFAYSFSIILIEIQDTIKSPPAEHK--TMRKATTLSIAVTTVFYLLCGCMGYAAFG 1008
Query: 269 NSVDDNIL--LSLEKPRWLIIAANIFVVVHVVGSYQVYAVPVFHMLESFLAE---KMNFK 323
++ N+L P WL+ AN+ +V+H+VG+YQV++ P+F +E + K NF
Sbjct: 1009DNAPGNLLTGFGFYNPYWLLDIANLAIVIHLVGAYQVFSQPLFAFVEKWSVRKWPKSNFV 1188
Query: 324 PS--------------RFLRFAIRNLYVSITMVLAITFPFFGGLLSFFGGFVFAPTTYFL 369
+ F R R ++V +T ++A+ PFF ++ G F F P T +
Sbjct: 1189TAEYDIPIPCFGVYQLNFFRLVWRTIFVLLTTLIAMLMPFFNDVVGILGAFGFWPLTVYF 1368
Query: 370 PCIMWIFIYKPKLFSLSWCANWV 392
P M+I K W + W+
Sbjct: 1369PIDMYISQKKIG----RWTSRWI 1425
>TC220754 similar to UP|Q85V22 (Q85V22) Histidine amino acid transporter,
partial (27%)
Length = 664
Score = 149 bits (376), Expect = 2e-36
Identities = 67/112 (59%), Positives = 85/112 (75%), Gaps = 11/112 (9%)
Frame = +1
Query: 303 VYAVPVFHMLESFLAEKMNFKPSRFLRFAIRNLYVSITMVLAITFPFFGGLLSFFGGFVF 362
+YA+PVF M+E+ + +K+NF+PSR LRF +RN+YV+ TM +AITFPFF GLL FFGGF F
Sbjct: 16 IYAMPVFDMIETVMVKKLNFEPSRMLRFVVRNVYVAFTMFIAITFPFFDGLLGFFGGFAF 195
Query: 363 APTTYFLPCIMWIFIYKPKLFSLSWCANWV-----------SPIGALRQVIL 403
APTTYFLPCIMW+ I+KPK +SLSW NW+ SPIG LR +I+
Sbjct: 196 APTTYFLPCIMWLAIHKPKRYSLSWFINWICIVLGLCLMILSPIGGLRTIII 351
>TC230068 similar to UP|Q85V22 (Q85V22) Histidine amino acid transporter,
partial (22%)
Length = 682
Score = 124 bits (310), Expect = 1e-28
Identities = 56/87 (64%), Positives = 66/87 (75%), Gaps = 11/87 (12%)
Frame = +3
Query: 328 LRFAIRNLYVSITMVLAITFPFFGGLLSFFGGFVFAPTTYFLPCIMWIFIYKPKLFSLSW 387
LRF +RNLYV+ TM +AITFPFFGGLL FFGGF FAPTTYFLPC+MW+ IYKP+ FS+SW
Sbjct: 3 LRFIVRNLYVAFTMFVAITFPFFGGLLGFFGGFAFAPTTYFLPCVMWLAIYKPRRFSMSW 182
Query: 388 CANWV-----------SPIGALRQVIL 403
ANW+ SPIG LR +I+
Sbjct: 183 WANWICIVFGLLLMILSPIGGLRSIII 263
>TC218569 similar to UP|Q93X14 (Q93X14) Amino acid permease AAP3, partial
(51%)
Length = 1108
Score = 109 bits (272), Expect = 3e-24
Identities = 68/207 (32%), Positives = 102/207 (48%), Gaps = 16/207 (7%)
Frame = +3
Query: 197 TKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWRGMIIAYLVVALCY 256
T+A ++G+F ALG+IAF Y+ V+LEIQ TI S P + +M + ++ V Y
Sbjct: 30 TEAQKVWGVFQALGNIAFAYSYSFVLLEIQDTIKSPPSEVK--TMKKAAKLSIAVTTTFY 203
Query: 257 FPVTIFGYRAFGNSVDDNIL--LSLEKPRWLIIAANIFVVVHVVGSYQVYAVPVFHMLES 314
GY AFG+S N+L K WLI AN +V+H+VG+YQVYA P+F +E
Sbjct: 204 MLCGCVGYAAFGDSAPGNLLAGFGFHKLYWLIDIANAAIVIHLVGAYQVYAQPLFAFVEK 383
Query: 315 FLAEK--------------MNFKPSRFLRFAIRNLYVSITMVLAITFPFFGGLLSFFGGF 360
A++ + R ++V IT V+++ PFF +L G
Sbjct: 384 ETAKRWPKIDKEFQISIPGLQSYNQNIFSLVWRTVFVIITTVISMLLPFFNDILGVIGAL 563
Query: 361 VFAPTTYFLPCIMWIFIYKPKLFSLSW 387
F P T + P M+I + +S+ W
Sbjct: 564 GFWPLTVYFPVEMYILQKRIPKWSMRW 644
>TC208738 similar to UP|Q7Y076 (Q7Y076) Amino acid permease 6, partial (52%)
Length = 966
Score = 100 bits (248), Expect = 2e-21
Identities = 73/232 (31%), Positives = 118/232 (50%), Gaps = 19/232 (8%)
Frame = +3
Query: 21 HNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMHES---VSGKRFD 77
H +TA++G+ VL +A++Q+GW G +L T +T+ + + + S V GKR
Sbjct: 273 HIITAVIGSGVLSLAWAIAQMGWVAGPAVLFAFSFITYFTSTLLADCYRSPDPVHGKRNY 452
Query: 78 KYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILC-------DDC 130
Y ++ + G R + Q + + VG+ I Y + + S+ + C D C
Sbjct: 453 TYSDVVRSVLGGRKFQLCGLAQYINL-VGVTIGYTITASISMVAVKRSNCFHKHGHHDKC 629
Query: 131 EPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASIH-------- 182
F++LFA +Q VLS +P+F+ + +S+VAA MS +YS+I S+
Sbjct: 630 YTSNNP-FMILFACIQIVLSQIPNFHKLWWLSIVAAVMSFAYSSIGLGLSVAKVAGGGEP 806
Query: 183 -RGALPDVQYSSRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTP 233
R L VQ T + ++ F A+GDIAF YA NV++EIQ T+ S+P
Sbjct: 807 VRTTLTGVQVGVDV-TGSEKVWRTFQAIGDIAFAYAYSNVLIEIQDTLKSSP 959
>AW397326
Length = 426
Score = 100 bits (248), Expect = 2e-21
Identities = 50/133 (37%), Positives = 80/133 (59%), Gaps = 7/133 (5%)
Frame = +3
Query: 196 STKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWRGMIIAYLVVALC 255
S + ++F NALG IAF + GHN+ LEIQST+PST + P++V MW+G +AY +A+C
Sbjct: 24 SQPSASVFLAMNALGIIAFSFRGHNLALEIQSTMPSTFKHPARVPMWKGAKVAYFFIAMC 203
Query: 256 YFPVTIFGYRAFGNSVDDNILL-------SLEKPRWLIIAANIFVVVHVVGSYQVYAVPV 308
FP+ I G+ A+GN + +L S + R ++ A + VV + + S+Q+Y++P
Sbjct: 204 LFPIAIGGFWAYGNQMPPGGILTALYAFHSHDISRGILALAFLLVVFNCLSSFQIYSMPA 383
Query: 309 FHMLESFLAEKMN 321
F E+ + N
Sbjct: 384 FDSFEAGYTSRTN 422
>TC222656
Length = 415
Score = 99.4 bits (246), Expect = 3e-21
Identities = 48/134 (35%), Positives = 80/134 (58%), Gaps = 4/134 (2%)
Frame = +2
Query: 101 LMVEVGIDIVYMVIGAKSLKKLHEILCDD---CEPIKTTYFIVLFAFVQYVLSHLPSFNS 157
L + G ++IG + + ++++C + +P+ T + ++F V VLS +P+ N+
Sbjct: 14 LYLSAGTCTTLIIIGGSTARTFYQVVCGETCTAKPMTTVEWYLVFTCVAVVLSQVPNLNT 193
Query: 158 VAGISLVAAAMSLSYSTIAWIASIHRGALPDVQYSS-RYSTKAGNIFGIFNALGDIAFGY 216
+AG++L+ AA + Y T W+ S+ RGALPDV Y+ R + F + NAL IAF +
Sbjct: 194 IAGVTLIGAATAEGYCTAIWVTSVARGALPDVSYNPVRTGNSVEDAFSVLNALXIIAFAF 373
Query: 217 AGHNVILEIQSTIP 230
GHN++LEIQS +P
Sbjct: 374 RGHNLMLEIQSAMP 415
>TC218082 similar to UP|P92934 (P92934) Amino acid permease 6, partial (40%)
Length = 638
Score = 89.0 bits (219), Expect = 4e-18
Identities = 55/153 (35%), Positives = 73/153 (46%), Gaps = 19/153 (12%)
Frame = +1
Query: 254 LCYFPVTIFGYRAFGNSVDDNIL--LSLEKPRWLIIAANIFVVVHVVGSYQVYAVPVFHM 311
L Y GY AFGN N L +P WLI ANI + VH+VG+YQV+ P+F
Sbjct: 10 LFYVLCGCLGYAAFGNDAPGNFLTGFGFYEPFWLIDFANICIAVHLVGAYQVFCQPIFGF 189
Query: 312 LESFLAEK------------MNFK-----PSRFLRFAIRNLYVSITMVLAITFPFFGGLL 354
+E++ E+ +NF P F R R YV IT ++A+ FPFF L
Sbjct: 190 VENWGKERWPNSQFVNGEHALNFPLCGTFPVNFFRVVWRTTYVIITALIAMMFPFFNDFL 369
Query: 355 SFFGGFVFAPTTYFLPCIMWIFIYKPKLFSLSW 387
G F P T + P M+I K + FS +W
Sbjct: 370 GLIGSLSFWPLTVYFPIEMYIKQSKMQRFSFTW 468
>TC220062 weakly similar to UP|Q93X13 (Q93X13) Amino acid permease AAP4,
partial (29%)
Length = 818
Score = 88.2 bits (217), Expect = 6e-18
Identities = 55/176 (31%), Positives = 83/176 (46%), Gaps = 19/176 (10%)
Frame = +2
Query: 236 PSKVSMWRGMIIAYLVVALCYFPVTIFGYRAFGNSVDDNILLSL--EKPRWLIIAANIFV 293
P +M + +IA V Y Y AFG++ N+L K WL+ AN +
Sbjct: 2 PENQTMKKASVIAVSVTTFLYXSCGGXXYAAFGDNTPGNLLTGFVSSKSYWLVNFANACI 181
Query: 294 VVHVVGSYQVYAVPVFHMLESFL--------------AEKMNFKPS---RFLRFAIRNLY 336
VVH+VGSYQVY+ P+F +E++ K+ P+ FL + R Y
Sbjct: 182 VVHLVGSYQVYSQPLFGTVENWFRFRFPDSEFVNHTYILKLPLLPAFELNFLSLSFRTAY 361
Query: 337 VSITMVLAITFPFFGGLLSFFGGFVFAPTTYFLPCIMWIFIYKPKLFSLSWCANWV 392
V+ T V+A+ FP+F +L G +F P T + P + IY + ++SW WV
Sbjct: 362 VASTTVIAMIFPYFNQILGVLGSIIFWPLTIYFP----VEIYLSQSSTVSWTTKWV 517
>TC203584 weakly similar to UP|Q93X15 (Q93X15) Amino acid permease AAP1,
partial (34%)
Length = 953
Score = 85.1 bits (209), Expect = 5e-17
Identities = 53/179 (29%), Positives = 80/179 (44%), Gaps = 19/179 (10%)
Frame = +2
Query: 228 TIPSTPEKPSKVSMWRGMIIAYLVVALCYFPVTIFGYRAFGNSVDDNILLSLE--KPRWL 285
T+ S P P +M + ++A + Y FGY AFGN N+L +P WL
Sbjct: 5 TLESPP--PENQTMKKASMVAIFITTFFYLCCGCFGYAAFGNDTPGNLLTGFGFFEPFWL 178
Query: 286 IIAANIFVVVHVVGSYQVYAVPVFHMLESFLAEKM-------NFKPSRF----------L 328
I AN +++H+VG YQ+Y+ P++ ++ + + K NF +
Sbjct: 179 IDLANACIILHLVGGYQIYSQPIYSTVDRWASRKFPNSGFVNNFYRVKLPLLPGFQLNLF 358
Query: 329 RFAIRNLYVSITMVLAITFPFFGGLLSFFGGFVFAPTTYFLPCIMWIFIYKPKLFSLSW 387
RF R YV T+ LAI FP+F +L G F P + P M+ K +S W
Sbjct: 359 RFCFRTTYVISTIGLAIFFPYFNQILGVLGAINFWPLAIYFPVEMYFVQQKIAAWSSKW 535
>TC222161 weakly similar to UP|Q9SGD7 (Q9SGD7) T23G18.9, partial (52%)
Length = 978
Score = 77.0 bits (188), Expect(2) = 6e-17
Identities = 54/169 (31%), Positives = 74/169 (42%), Gaps = 10/169 (5%)
Frame = +1
Query: 229 IPSTPEKPSKVSMWRGMIIAYLVVALCYFPVTIFGYRAFGNSVDDNILLSLEK------P 282
I +T P K M R + Y+VV +F V I GY AFGN + I S P
Sbjct: 220 IQATLAPPVKGKMLRSLCACYVVVLFSFFCVAISGYWAFGNQAEGLIFSSFVDSNKPLAP 399
Query: 283 RWLIIAANIFVVVHVVGSYQVYAVPVFHMLESFLA--EKMNFKPSRFL-RFAIRNLYVSI 339
+WLI NI + + + Y P +LE E F P + R R+L V
Sbjct: 400 KWLIYMPNICTIAQLTANGVEYLQPTNVILEQIFGDPEIPEFSPRNVIPRLISRSLAVIT 579
Query: 340 TMVLAITFPFFGGLLSFFGGFVFAPTTYFLPCIMWIFIYKP-KLFSLSW 387
++A PFFG + S G F + P + LP I + +KP K S+ W
Sbjct: 580 ATIIAAMLPFFGDMNSLIGAFGYMPLDFILPMIFFNMTFKPSKRSSIFW 726
Score = 28.1 bits (61), Expect(2) = 6e-17
Identities = 18/40 (45%), Positives = 23/40 (57%)
Frame = +3
Query: 187 PDVQYSSRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQ 226
P+ YS + T +FGIFNA+ IA Y G +I EIQ
Sbjct: 18 PEKDYSLKGDT-TNRLFGIFNAIPIIATTY-GSGIIPEIQ 131
>TC232997 weakly similar to UP|Q9ARG2 (Q9ARG2) Amino acid transporter,
partial (18%)
Length = 487
Score = 81.6 bits (200), Expect = 6e-16
Identities = 45/142 (31%), Positives = 67/142 (46%), Gaps = 16/142 (11%)
Frame = +3
Query: 263 GYRAFGNSVDDNILLSLEKPRWLIIAANIFVVVHVVGSYQVYAVPVFHMLE--------- 313
GY AFG++ NIL +P WL+ N F+V+H++G+YQV P F ++E
Sbjct: 27 GYAAFGDNTPGNILTGFTEPFWLVALGNGFIVIHMIGAYQVMGQPFFRIVEIGANIAWPN 206
Query: 314 -SFLAEKMNFKPS------RFLRFAIRNLYVSITMVLAITFPFFGGLLSFFGGFVFAPTT 366
F+ ++ F R R ++V + +LA+ PFF +LS G F P
Sbjct: 207 SDFINKEYPFIVGGLMVRFNLFRLVWRTIFVILATILAMVMPFFSEVLSLLGAIGFGPLV 386
Query: 367 YFLPCIMWIFIYKPKLFSLSWC 388
F+P M I + SL WC
Sbjct: 387 VFIPIQMHIAQKSIRKLSLRWC 452
>TC231343 similar to UP|Q9FF99 (Q9FF99) Amino acid transporter, partial (27%)
Length = 700
Score = 75.5 bits (184), Expect = 4e-14
Identities = 60/223 (26%), Positives = 104/223 (45%), Gaps = 19/223 (8%)
Frame = +3
Query: 21 HNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMHE--------SVS 72
H VT ++G+ VL P++ +QLGW G ++L TL++++ + + + S
Sbjct: 48 HIVTGVIGSGVLSLPWSTAQLGWLAGPFSILLIASTTLFSSFLLCNTYRHPHPEYGPNRS 227
Query: 73 GKRFDKYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILCDD--- 129
D H + G GL + + G I +++ A SL+ + C
Sbjct: 228 ASYLDVVHLHLGISNGRLSGLLVSISLY-----GFAIAFVITTAISLRTIQNSFCYHNKG 392
Query: 130 ----CEPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIAS----I 181
CE + Y+++LF +Q VLS +P+F+++ +S+VAA MS +YS I S I
Sbjct: 393 PEAACESVDA-YYMLLFGAIQIVLSQIPNFHNIKWLSVVAAIMSFTYSFIGMGLSIAQII 569
Query: 182 HRGALPDVQYSSRYSTKAGNIFGIFNALGDIAFGYAGHNVILE 224
+G S A ++ + ALGDI+F Y +++E
Sbjct: 570 EKGHAEGSIGGISTSNGAEKLWLVSQALGDISFSYPFSTILME 698
>TC203550 weakly similar to UP|Q9SXF7 (Q9SXF7) Amino acid transporter-like
protein 1, partial (21%)
Length = 1426
Score = 71.6 bits (174), Expect = 6e-13
Identities = 46/157 (29%), Positives = 74/157 (46%), Gaps = 9/157 (5%)
Frame = +3
Query: 256 YFPVTIFGYRAFGNSVDDN--ILLSL------EKPRWLIIAANIFVVVHVVGSYQVYAVP 307
YFP + A+G + N +L +L + R+++ + FVVV+ + S+Q+Y +P
Sbjct: 18 YFPWPLEAIXAYGQLIPANGGMLTALYQFHSRDVSRFVLGLTSFFVVVNGLCSFQIYGMP 197
Query: 308 VFHMLESFLAEKMNFKPSRFLRFAIRNLYVSITMVLAITFPFFGGLLSFFGGFVFAPTTY 367
F +ES +M +LR IR + + + + PF + GG V P T+
Sbjct: 198 AFDDMESGYTTRMKKPCPWWLRAFIRVFFGFLCFFIGVAVPFLSQMAGLIGG-VALPVTF 374
Query: 368 FLPCIMWIFIYKPKLFSLSWCANW-VSPIGALRQVIL 403
PC MW+ KPK +S W NW + +G IL
Sbjct: 375 AYPCFMWLKTKKPKKYSAMWWLNWFLGTLGVALSAIL 485
>TC229508 similar to UP|Q941E6 (Q941E6) AT5g23810/MRO11_15, partial (39%)
Length = 865
Score = 60.5 bits (145), Expect(2) = 6e-13
Identities = 35/124 (28%), Positives = 57/124 (45%), Gaps = 17/124 (13%)
Frame = +1
Query: 281 KPRWLIIAANIFVVVHVVGSYQVYAVPVFHMLESFLAEKM-------NFKPSR------- 326
+P WLI AN +V+H+VG YQ+Y+ P++ ++ + +++ NF +
Sbjct: 124 EPYWLIDFANACIVLHLVGGYQIYSQPIYGAVDRWCSKRYPNSGFVNNFYQLKLPRLPAF 303
Query: 327 ---FLRFAIRNLYVSITMVLAITFPFFGGLLSFFGGFVFAPTTYFLPCIMWIFIYKPKLF 383
R R YV T LAI FP+F ++ G F P + P M+ K + +
Sbjct: 304 QLNMFRICFRTAYVVSTTGLAILFPYFNQVIGVLGALGFWPLAIYFPVEMYFVQRKVEAW 483
Query: 384 SLSW 387
S W
Sbjct: 484 SRKW 495
Score = 31.2 bits (69), Expect(2) = 6e-13
Identities = 14/38 (36%), Positives = 19/38 (49%)
Frame = +2
Query: 243 RGMIIAYLVVALCYFPVTIFGYRAFGNSVDDNILLSLE 280
+ +IA L+ Y FGY AFGN N+L L+
Sbjct: 5 KASMIAILITTFFYLCCGCFGYAAFGNQTPGNLLTGLD 118
>TC203503 weakly similar to UP|Q9SXF7 (Q9SXF7) Amino acid transporter-like
protein 1, partial (21%)
Length = 713
Score = 70.5 bits (171), Expect = 1e-12
Identities = 41/127 (32%), Positives = 62/127 (48%), Gaps = 1/127 (0%)
Frame = +2
Query: 278 SLEKPRWLIIAANIFVVVHVVGSYQVYAVPVFHMLESFLAEKMNFKPSRFLRFAIRNLYV 337
S + R+++ + FVVV+ + S+Q+Y +P F +ES +M +LR IR +
Sbjct: 32 SRDVSRFVLGLTSFFVVVNGLCSFQIYGMPAFDDMESGYTARMKKPCPWWLRAFIRVFFG 211
Query: 338 SITMVLAITFPFFGGLLSFFGGFVFAPTTYFLPCIMWIFIYKPKLFSLSWCANW-VSPIG 396
+ + + PF L GG V P T+ PC MW+ KPK SL W NW + +G
Sbjct: 212 FLCFFIGVAVPFLSQLAGLIGG-VALPVTFAYPCFMWLKTKKPKKLSLMWWLNWFLGTLG 388
Query: 397 ALRQVIL 403
IL
Sbjct: 389 VALSAIL 409
>TC203853 weakly similar to UP|Q9SXF7 (Q9SXF7) Amino acid transporter-like
protein 1, partial (21%)
Length = 904
Score = 68.2 bits (165), Expect = 6e-12
Identities = 32/84 (38%), Positives = 50/84 (59%)
Frame = +2
Query: 9 KSRNAKCCYSAFHNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMH 68
+SRN Y+AFH + + +G L P A + LGW G L L+++ LY + ++++H
Sbjct: 443 ESRNGNAYYAAFHILNSNIGFQALMLPVAFATLGWAWGTVCLSLAFVWQLYAIFLLVQLH 622
Query: 69 ESVSGKRFDKYHELSQHAFGERLG 92
ESV G R +Y L+ AFG++LG
Sbjct: 623 ESVPGIRHSRYLFLAMAAFGKKLG 694
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.329 0.141 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,120,278
Number of Sequences: 63676
Number of extensions: 302595
Number of successful extensions: 2354
Number of sequences better than 10.0: 100
Number of HSP's better than 10.0 without gapping: 2311
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2328
length of query: 403
length of database: 12,639,632
effective HSP length: 99
effective length of query: 304
effective length of database: 6,335,708
effective search space: 1926055232
effective search space used: 1926055232
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC135505.15


