

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135464.1 - phase: 0 /pseudo
(317 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
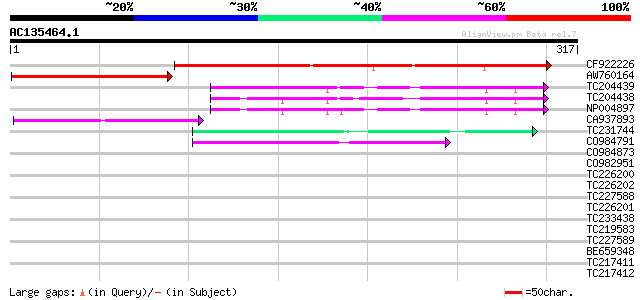
Score E
Sequences producing significant alignments: (bits) Value
CF922226 189 2e-48
AW760164 similar to GP|11994422|dbj oxidoreductase short-chain ... 87 8e-18
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 53 2e-07
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 51 8e-07
NP004897 gag-protease polyprotein 50 2e-06
CA937893 similar to GP|20805072|dbj retrovirus-related pol polyp... 47 1e-05
TC231744 44 1e-04
CO984791 42 3e-04
CO984873 40 0.002
CO982951 39 0.003
TC226200 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 38 0.005
TC226202 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 37 0.009
TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L1... 37 0.015
TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 36 0.020
TC233438 similar to UP|Q6XD84 (Q6XD84) SR RNA-binding protein, p... 34 0.076
TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich p... 34 0.076
TC227589 33 0.17
BE659348 weakly similar to PIR|JC7809|JC78 sulfakinin receptor p... 33 0.17
TC217411 similar to UP|Q8L7S8 (Q8L7S8) At5g26740, partial (26%) 33 0.17
TC217412 similar to UP|Q6L724 (Q6L724) ATP-dependent RNA helicas... 33 0.22
>CF922226
Length = 667
Score = 189 bits (479), Expect = 2e-48
Identities = 98/219 (44%), Positives = 144/219 (65%), Gaps = 8/219 (3%)
Frame = -3
Query: 93 MTKSLAHRQLLKQQLYSFKMMESKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLCSL 152
MTKSL +R KQ LYSFKM E +S+ EQL FNK++ DL NI+V ++DED+ALLLLC L
Sbjct: 665 MTKSLVNRLYXKQSLYSFKMHEDRSVGEQLDLFNKLILDLENIDVTIDDEDQALLLLCYL 486
Query: 153 PKSFEHFKDTILYGKEGTATLEEVQAALRTKELTKFKDMKVDEGSEGLNV---TRGRNEH 209
PKS+ HFK+T+L+G++ + +L+EVQ AL +KEL + K+ K EGL T ++
Sbjct: 485 PKSYSHFKETLLFGRD-SVSLDEVQTALNSKELNERKEKKSSASGEGLTARGKTFKKDSE 309
Query: 210 RGKGKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASN----- 264
K K K ++ +G + K +C+ C K+GH +K CP++ +G + + ++ N
Sbjct: 308 FDKKKQKPENQKNGEG-NIFKIRCYHCKKEGHTRKVCPERQKNGGSNNRKKDSGNAAIVQ 132
Query: 265 EEGYESAGALVVTSWEPEKSWVLDSGCSYHMCPRKEYFE 303
++GYESA AL+V+ PE W++DSGCS+HM P K +FE
Sbjct: 131 DDGYESAEALMVSEKNPETKWIMDSGCSWHMTPNKSWFE 15
>AW760164 similar to GP|11994422|dbj oxidoreductase short-chain
dehydrogenase/reductase family-like protein {Arabidopsis
thaliana}, partial (9%)
Length = 428
Score = 87.4 bits (215), Expect = 8e-18
Identities = 46/91 (50%), Positives = 65/91 (70%), Gaps = 1/91 (1%)
Frame = +3
Query: 2 MGS-KWDIEKFTGSNDFGL*KVKMQAILTQQKCVEALKGEAEMPATLTQEEKREMVDKAK 60
MGS K+++EKFTG NDFGL +KM+A+L QQ VEAL GE ++ + +K+ ++ KA
Sbjct: 75 MGSAKYEVEKFTGQNDFGLC*LKMRALLVQQGLVEALDGEIKLEKMMADGDKKALLQKAY 254
Query: 61 SAIVLCLGDKVLRDVAREATAASMLAKWESL 91
+AI+L LGDKVLR V++E TA + +K E L
Sbjct: 255 NAIILSLGDKVLRQVSKETTAVGVWSKLEVL 347
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 53.1 bits (126), Expect = 2e-07
Identities = 54/213 (25%), Positives = 92/213 (42%), Gaps = 24/213 (11%)
Frame = +1
Query: 113 MESKSISEQLTEFNKILDDL-ANIEVNMEDEDKALLLLCSLPKSFEHFKDTILYGKEGTA 171
++S+ I +Q + K++ DL A E + E+ + + L E+ +I +G+
Sbjct: 1135 IKSEKILQQEAQLKKVIADLEAEKEAHKEEISELKGEVGFLNSKLENMTKSIKMLNKGSD 1314
Query: 172 TLEEV-----------------QAALRTKELTKFKDMKVDEGSEGLNVTRGRNEHRGKGK 214
TL+EV ++A RT +T+F K G+ ++HR +
Sbjct: 1315 TLDEVLLLGKNAGNQRGLGFNPKSAGRTT-MTEFVPAKNRTGAT-------MSQHRSRHH 1470
Query: 215 GKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASNE----EGYES 270
G + KS+ K K++C C K GH K C G Q + + + +++
Sbjct: 1471 GMQQKKSKRK-----KWRCHYCGKYGHIKPFCYHLHGHPHHGTQSSNSRKKMMWVPKHKA 1635
Query: 271 AGALVVTSWEP--EKSWVLDSGCSYHMCPRKEY 301
+V TS ++ W LDSGCS HM KE+
Sbjct: 1636 VSLVVHTSLRASAKEDWYLDSGCSRHMTGVKEF 1734
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 50.8 bits (120), Expect = 8e-07
Identities = 54/216 (25%), Positives = 91/216 (42%), Gaps = 27/216 (12%)
Frame = +1
Query: 113 MESKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLCS----LPKSFEHFKDTILYGKE 168
++S+ I +Q + K++ AN+E E ++ + L L E+ +I +
Sbjct: 1138 IKSEKILQQEAQLKKVI---ANLEAEKEAHEEEISELKGEVGFLNSKLENMTKSIKMLNK 1308
Query: 169 GTATLEEV-----------------QAALRTKELTKFKDMKVDEGSEGLNVTRGRNEHRG 211
G+ L+EV ++A RT +T+F K S G +++ R+ H G
Sbjct: 1309 GSDMLDEVLQLGKNVGNQRGLGFNHKSAGRTT-MTEFVPAK---NSTGATMSQHRSRHHG 1476
Query: 212 KGKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASNE----EG 267
+ KS+ K K++C C K GH K C G Q + + +
Sbjct: 1477 TQQKKSKRK---------KWRCHYCGKYGHIKPFCYHLHGHPHHGTQSSSSGRKMMWVPK 1629
Query: 268 YESAGALVVTSWEP--EKSWVLDSGCSYHMCPRKEY 301
++ +V TS ++ W LDSGCS HM KE+
Sbjct: 1630 HKIVSLVVHTSLRASAKEDWYLDSGCSRHMTGVKEF 1737
>NP004897 gag-protease polyprotein
Length = 1923
Score = 49.7 bits (117), Expect = 2e-06
Identities = 52/215 (24%), Positives = 88/215 (40%), Gaps = 26/215 (12%)
Frame = +1
Query: 113 MESKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLCS----LPKSFEHFKDTILYGKE 168
++S+ I +Q + K++ AN+E E ++ + L L E+ +I +
Sbjct: 1138 IKSEKILQQEAQLKKVI---ANLEAEKEAHEEEISELKGEVGFLNSKLENMTKSIKMLNK 1308
Query: 169 GTATLEEV---------QAALRTKE-------LTKFKDMKVDEGSEGLNVTRGRNEHRGK 212
G+ L+EV Q L +T+F K+ G+ ++HR +
Sbjct: 1309 GSDMLDEVLQLGKNVGNQRGLGFNHKSAGRITMTEFVPAKISTGAT-------MSQHRSR 1467
Query: 213 GKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASNE----EGY 268
G + KS+ K K++C C K GH K C G Q + + + +
Sbjct: 1468 HHGTQQKKSKRK-----KWRCHYCGKYGHIKPFCYHLHGHPHHGTQSSSSRRKMMWVPKH 1632
Query: 269 ESAGALVVTSWEP--EKSWVLDSGCSYHMCPRKEY 301
+ +V TS ++ W LDSGCS HM KE+
Sbjct: 1633 KIVSLVVHTSLRASAKEDWYLDSGCSRHMTGVKEF 1737
>CA937893 similar to GP|20805072|dbj retrovirus-related pol polyprotein from
transposon TNT 1-94-like, partial (7%)
Length = 412
Score = 47.0 bits (110), Expect = 1e-05
Identities = 31/106 (29%), Positives = 55/106 (51%)
Frame = -2
Query: 3 GSKWDIEKFTGSNDFGL*KVKMQAILTQQKCVEALKGEAEMPATLTQEEKREMVDKAKSA 62
G+K+++ KF G+ +F L + +++ +L Q ++ L+ E E+ ++ +
Sbjct: 315 GAKFEVGKFDGTGNFRLWQKRVKDLLA*QGLLKVLRDSKSNNTEALDWE--EL*ERTATT 142
Query: 63 IVLCLGDKVLRDVAREATAASMLAKWESLYMTKSLAHRQLLKQQLY 108
I LCL D+ L + A + K ES YM KSL ++ L Q+LY
Sbjct: 141 IRLCLVDEFLYHMMELAFPGEVWKKLESQYMLKSLTNKLYLMQKLY 4
>TC231744
Length = 794
Score = 43.9 bits (102), Expect = 1e-04
Identities = 42/194 (21%), Positives = 78/194 (39%), Gaps = 1/194 (0%)
Frame = +2
Query: 103 LKQQLYSFKMMESKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLCSLPKSFEHFKDT 162
L +L S K +I E + E + + L ++++ + ++ L+L SLP F FK +
Sbjct: 41 LLDKLISMKYKGKGNIREYIMEISNLASKLKSLKLELGEDLFVHLVLISLPAHFGQFKVS 220
Query: 163 ILYGKEGTATLEEVQAALRTKELTKFKDMKVDEGSEGLNVTRGRNEHRGKGKGKSRSKSR 222
K+ + E + ++ +E +D N+ R K K + S
Sbjct: 221 YNTQKDKWSLNELISHCVQEEERL*-RD----------RTESAHNKKRKKTKDVAEKTS* 367
Query: 223 SKGFDKSK-YKCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASNEEGYESAGALVVTSWEP 281
K K + + C+ C K H KK C P ++ + V ++ P
Sbjct: 368 QKKQQKDEEFTCYFCKKSRHMKKKC--------PKYAAWRVKKDKFLTLVCSEVNLAFVP 523
Query: 282 EKSWVLDSGCSYHM 295
+ +W +DSG + H+
Sbjct: 524 KDTWWVDSGATTHI 565
>CO984791
Length = 672
Score = 42.4 bits (98), Expect = 3e-04
Identities = 34/145 (23%), Positives = 65/145 (44%), Gaps = 1/145 (0%)
Frame = -3
Query: 103 LKQQLYSFKMMESKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLCSLPKSFEHFKDT 162
L +L S K +I E + E + + L ++++ + ++ L+L SLP F FK +
Sbjct: 607 LLAKLISMKYKGKGNIREYIIEMSNLASKLKSLKLELGEDLLMYLVLISLPAHFGQFKVS 428
Query: 163 ILYGKEGTATLEEVQAALRTKELTKFKDMKVDEGSEGLNVTRGRNEHRGKGKGKSRSKSR 222
K+ + E + ++ +E + D ++ +N+ K KG + S
Sbjct: 427 YNT*KDKWSLNELISHCVQEEE-----SL*RDRTESAHLISTSQNKKWKKTKGAA*GTS* 263
Query: 223 SKGFDKSK-YKCFLCHKQGHFKKDC 246
K +K + + C+ K GH KK+C
Sbjct: 262 QKKQNKDEEFACYFYKKSGHMKKEC 188
>CO984873
Length = 754
Score = 39.7 bits (91), Expect = 0.002
Identities = 15/33 (45%), Positives = 23/33 (69%)
Frame = -2
Query: 214 KGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDC 246
+ K++ K +K K++ KCF C+K+GH KKDC
Sbjct: 414 QAKNKGKITAKPVIKNESKCFFCNKKGHIKKDC 316
>CO982951
Length = 757
Score = 38.9 bits (89), Expect = 0.003
Identities = 18/41 (43%), Positives = 22/41 (52%), Gaps = 3/41 (7%)
Frame = -3
Query: 212 KGKGKSRSKSRSKGFDKSKYK---CFLCHKQGHFKKDCPDK 249
KGKG + + SK K K C C K GHF+KDCP +
Sbjct: 296 KGKGPLKINNNSKQIQKKTSKRNNCHFCGKSGHFQKDCPKR 174
>TC226200 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (89%)
Length = 936
Score = 38.1 bits (87), Expect = 0.005
Identities = 21/56 (37%), Positives = 28/56 (49%)
Frame = +2
Query: 198 EGLNVTRGRNEHRGKGKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGGDG 253
+G N R H +G G R RS G D KC+ C + GHF ++C +GG G
Sbjct: 245 DGKNGWRVELSHNSRGGGGGRG-GRSGGSD---LKCYECGEPGHFARECRMRGGSG 400
>TC226202 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (72%)
Length = 1003
Score = 37.4 bits (85), Expect = 0.009
Identities = 21/55 (38%), Positives = 27/55 (48%)
Frame = +3
Query: 199 GLNVTRGRNEHRGKGKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGGDG 253
G N R H +G G R RS G D KC+ C + GHF ++C +GG G
Sbjct: 744 GKNGWRVELSHNSRGGGGGRG-GRSGGSD---LKCYECGEPGHFARECRMRGGSG 896
>TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L16.11 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(10%)
Length = 1300
Score = 36.6 bits (83), Expect = 0.015
Identities = 14/37 (37%), Positives = 20/37 (53%)
Frame = +1
Query: 233 CFLCHKQGHFKKDCPDKGGDGSPSVQVAEASNEEGYE 269
CF+C K GH KDCP+K S S+ + G++
Sbjct: 184 CFICKKGGHRAKDCPEKHTSTSKSIAICLKCGNSGHD 294
>TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (72%)
Length = 820
Score = 36.2 bits (82), Expect = 0.020
Identities = 19/50 (38%), Positives = 26/50 (52%)
Frame = +1
Query: 204 RGRNEHRGKGKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGGDG 253
R + H +G G R RS G D KC+ C + GHF ++C +GG G
Sbjct: 172 RVKLSHNSRGGGGGRG-GRSGGSD---LKCYECGEPGHFARECRMRGGSG 309
>TC233438 similar to UP|Q6XD84 (Q6XD84) SR RNA-binding protein, partial (5%)
Length = 856
Score = 34.3 bits (77), Expect = 0.076
Identities = 20/63 (31%), Positives = 28/63 (43%)
Frame = +2
Query: 192 KVDEGSEGLNVTRGRNEHRGKGKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGG 251
K GS + +R R + KG+ KSR K S + + +G KKDC + G
Sbjct: 197 KSHRGSATDDTSRKREHAKDKGEHKSRQKDASNNDRHRRRRSSSVSSRGRTKKDCINHAG 376
Query: 252 DGS 254
D S
Sbjct: 377 DSS 385
>TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich protein 2,
partial (45%)
Length = 995
Score = 34.3 bits (77), Expect = 0.076
Identities = 26/90 (28%), Positives = 39/90 (42%), Gaps = 1/90 (1%)
Frame = +2
Query: 165 YGKEGTATLEEVQAALRTKELTKFKDMKVDEGSEGLNVTRGRNEHR-GKGKGKSRSKSRS 223
YG +G +V +A+R++ + G G RGR R G G+G+ R R
Sbjct: 257 YGDDGRTMAVDVTSAVRSR---------LPGGFRGGGGGRGRGGGRYGGGEGRGRGFGRR 409
Query: 224 KGFDKSKYKCFLCHKQGHFKKDCPDKGGDG 253
G + C+ C + GH +DC G G
Sbjct: 410 GGGPE----CYNCGRIGHLARDCYHGQGGG 487
Score = 33.1 bits (74), Expect = 0.17
Identities = 16/46 (34%), Positives = 21/46 (44%)
Frame = +2
Query: 205 GRNEHRGKGKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKG 250
GRN RG G G CF C ++GHF ++CP+ G
Sbjct: 500 GRNRRRGGGGGGGGG-------------CFNCGEEGHFARECPNVG 598
>TC227589
Length = 547
Score = 33.1 bits (74), Expect = 0.17
Identities = 14/37 (37%), Positives = 19/37 (50%)
Frame = +2
Query: 233 CFLCHKQGHFKKDCPDKGGDGSPSVQVAEASNEEGYE 269
CF+C K GH KDC +K S SV + G++
Sbjct: 170 CFICKKGGHRAKDCLEKHTSRSKSVAICLKCGNSGHD 280
>BE659348 weakly similar to PIR|JC7809|JC78 sulfakinin receptor protein
DSK-R1 - fruit fly (Drosophila melanogaster), partial
(6%)
Length = 770
Score = 33.1 bits (74), Expect = 0.17
Identities = 39/185 (21%), Positives = 64/185 (34%), Gaps = 28/185 (15%)
Frame = -1
Query: 146 LLLLCSLPKSFEHFKDTILYGKEGTATLEEVQAALRTKELTKFKDMKVDEGSEGLNVT-R 204
+L+L + F+H +D +L +E +LE + L K VD + V+ R
Sbjct: 620 VLVLRGMHPDFDHIRDQVLTSQE-VPSLENLITRLLRVPSPKIGGNSVDNIETSVMVSNR 444
Query: 205 GRNEHRGKGKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASN 264
G RG G++ R C C + GH + C G SV ++++
Sbjct: 443 GGQGGRGNQGGRAGRGGRP*--------CSYCKRVGHTQDTCYSIHGFPGKSVNISKSET 288
Query: 265 EE---------------------------GYESAGALVVTSWEPEKSWVLDSGCSYHMCP 297
E G+ S A + + W++DSG S H+
Sbjct: 287 SEIKFLEADYQEYLQLKATKESQTSSVISGHNST-ACISQVGNNQSPWIIDSGASDHIAS 111
Query: 298 RKEYF 302
F
Sbjct: 110 NSSLF 96
>TC217411 similar to UP|Q8L7S8 (Q8L7S8) At5g26740, partial (26%)
Length = 1275
Score = 33.1 bits (74), Expect = 0.17
Identities = 19/58 (32%), Positives = 24/58 (40%), Gaps = 8/58 (13%)
Frame = +3
Query: 199 GLNVTRGRNEHRGKGK--------GKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPD 248
G N G + R G+ G SR SR D+ CF C + GH DCP+
Sbjct: 690 GRNFKTGNSWSRAAGRSSGDDWLIGGSRRSSRPSSSDRFGGACFNCGESGHRASDCPN 863
>TC217412 similar to UP|Q6L724 (Q6L724) ATP-dependent RNA helicase, partial
(3%)
Length = 868
Score = 32.7 bits (73), Expect = 0.22
Identities = 19/58 (32%), Positives = 23/58 (38%), Gaps = 8/58 (13%)
Frame = +3
Query: 199 GLNVTRGRNEHRGKGK--------GKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPD 248
G N G + R GK G R SR D+ CF C + GH DCP+
Sbjct: 186 GRNFKTGNSWSRAAGKSSGDDWLIGGGRRSSRPSSSDRFGGTCFNCGESGHRASDCPN 359
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.132 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,964,510
Number of Sequences: 63676
Number of extensions: 157280
Number of successful extensions: 1183
Number of sequences better than 10.0: 77
Number of HSP's better than 10.0 without gapping: 1055
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1146
length of query: 317
length of database: 12,639,632
effective HSP length: 97
effective length of query: 220
effective length of database: 6,463,060
effective search space: 1421873200
effective search space used: 1421873200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC135464.1


