

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135461.15 + phase: 0 /pseudo
(1184 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
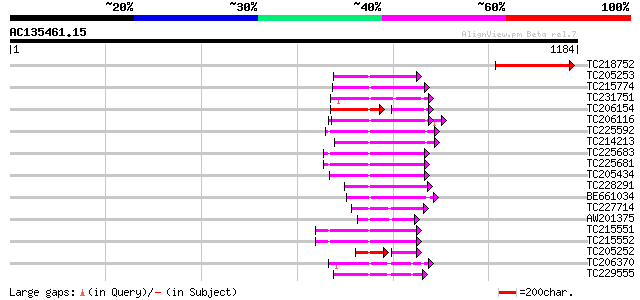
Score E
Sequences producing significant alignments: (bits) Value
TC218752 similar to UP|Q9M8Z0 (Q9M8Z0) T6K12.4 protein, partial ... 238 9e-63
TC205253 homologue to UP|O99018 (O99018) Chloroplast protease pr... 144 2e-34
TC215774 similar to UP|Q94ES0 (Q94ES0) AAA-metalloprotease FtsH,... 141 1e-33
TC231751 homologue to GB|BAB08420.1|9757998|AB025622 cell divisi... 135 8e-32
TC206154 homologue to UP|FTSH_MEDSA (Q9BAE0) Cell division prote... 88 3e-30
TC206116 UP|CC48_SOYBN (P54774) Cell division cycle protein 48 h... 124 3e-28
TC225592 homologue to GB|AAP78936.1|32306505|BT009668 At5g19990 ... 116 5e-26
TC214213 homologue to UP|PRSA_BRACM (O23894) 26S protease regula... 108 2e-23
TC225683 homologue to UP|Q9SZD4 (Q9SZD4) 26S proteasome subunit ... 105 2e-22
TC225681 homologue to UP|Q9SZD4 (Q9SZD4) 26S proteasome subunit ... 105 2e-22
TC205434 homologue to UP|PRS6_ARATH (Q9SEI4) 26S protease regula... 102 8e-22
TC228291 homologue to UP|Q9FXT8 (Q9FXT8) 26S proteasome regulato... 102 1e-21
BE661034 similar to SP|Q9ZPR1|C48B Cell division control protein... 102 1e-21
TC227714 similar to UP|C48C_ARATH (Q9SS94) Cell division control... 96 9e-20
AW201375 homologue to GP|17065470|gb cell division protein FtsH-... 93 6e-19
TC215551 homologue to UP|PRS7_SPIOL (Q41365) 26S protease regula... 93 8e-19
TC215552 UP|PRS7_ARATH (Q9SSB5) 26S protease regulatory subunit ... 93 8e-19
TC205252 homologue to UP|Q9ZP50 (Q9ZP50) FtsH-like protein Pftf ... 59 1e-18
TC206370 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, part... 91 4e-18
TC229555 similar to GB|AAM62497.1|21553404|AY085265 26S proteaso... 89 9e-18
>TC218752 similar to UP|Q9M8Z0 (Q9M8Z0) T6K12.4 protein, partial (13%)
Length = 916
Score = 238 bits (608), Expect = 9e-63
Identities = 125/165 (75%), Positives = 137/165 (82%)
Frame = +1
Query: 1014 CSFITKF*CCS*SLA*ASILAGNWMYENHESKK*WLH*WEYRIKIIS*KETCILFWFLCC 1073
CSFITKF* C *SLA* SILAGN MY+NH+SKK* H WE+RIKIIS*KE C+LFWFLCC
Sbjct: 1 CSFITKF**C**SLA*TSILAGNRMYKNHKSKK*RFHQWEFRIKIIS*KEACVLFWFLCC 180
Query: 1074 IPNVTSFWGRKLTVFI*DAAGTRDCYADGHSIWMGA***CCSLLL**CGCYIKHGR*P*V 1133
IPNVTSFWGRKLT +I*D AGTRD Y +GHS+W+G ** C++L **CG IKHG *P*+
Sbjct: 181 IPNVTSFWGRKLTFYI*DTAGTRDSYTNGHSVWVGT**QSCNILP**CGYGIKHGG*P*I 360
Query: 1134 CDDS*SSKDV*FGISQSKRNAAEKSPGTRKDCRGIT*I*NFD*KG 1178
CD S*S KDV*FGISQSKRN AEK P T KDC GIT +*NFD KG
Sbjct: 361 CDGS*S*KDV*FGISQSKRNPAEKPPCT*KDCGGITRV*NFDWKG 495
>TC205253 homologue to UP|O99018 (O99018) Chloroplast protease precursor,
partial (69%)
Length = 1678
Score = 144 bits (364), Expect = 2e-34
Identities = 79/184 (42%), Positives = 109/184 (58%)
Frame = +2
Query: 677 IPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAE 736
+ ++ + +D K++ EVV FL+ P F +GAR P+GVL+VG GTGKT LA AIA E
Sbjct: 50 VTFDDVAGVDEAKQDFMEVVEFLKKPERFTAVGARIPKGVLLVGPPGTGKTLLAKAIAGE 229
Query: 737 AKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTE 796
A VP I + M+VG AS VR+LF+ A++ AP I+FV++ D RG I
Sbjct: 230 AGVPFFSISGSEF-VEMFVGVGASRVRDLFRKAKENAPCIVFVDEIDAVGRQRGTGIGGG 406
Query: 797 NQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAER 856
N + E +NQLL E+DGFE G++++A T + +D AL RPGR DR + P R
Sbjct: 407 NDEREQTLNQLLTEMDGFEGNTGIIVIAATNRVDILDSALLRPGRFDRQVTVDVPDIRGR 586
Query: 857 ENIL 860
IL
Sbjct: 587 TEIL 598
>TC215774 similar to UP|Q94ES0 (Q94ES0) AAA-metalloprotease FtsH, partial
(79%)
Length = 2084
Score = 141 bits (356), Expect = 1e-33
Identities = 80/202 (39%), Positives = 112/202 (54%)
Frame = +3
Query: 674 KPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAI 733
K I + + D K+EI E V FL+NP+ ++E+GA+ P+G L+VG GTGKT LA A
Sbjct: 480 KNKIYFKDVAGCDEAKQEIMEFVHFLKNPKKYEELGAKIPKGALLVGPPGTGKTLLAKAT 659
Query: 734 AAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFI 793
A E+ VP + I M+VG S VR LFQ AR +P I+F+++ D R
Sbjct: 660 AGESGVPFLSISGSDFME-MFVGVGPSRVRNLFQEARQCSPSIVFIDEIDAIGRARRGSF 836
Query: 794 HTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQ 853
N + E+ +NQLLVE+DGF GVV++A T + +D+AL RPGR DR + +P
Sbjct: 837 SGANDERESTLNQLLVEMDGFGTTSGVVVLAGTNRPEILDKALLRPGRFDRQITIDKPDI 1016
Query: 854 AERENILYSAAKETMDDQLVEY 875
R+ I K+ D Y
Sbjct: 1017KGRDQIFQIYLKKIKLDHEPSY 1082
>TC231751 homologue to GB|BAB08420.1|9757998|AB025622 cell division protein
FtsH protease-like {Arabidopsis thaliana;} , partial
(30%)
Length = 733
Score = 135 bits (341), Expect = 8e-32
Identities = 84/223 (37%), Positives = 119/223 (52%), Gaps = 7/223 (3%)
Frame = +1
Query: 670 KRVKKPPIPLNNFSSI------DSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERG 723
K + K +P N + D K+E+ EVV +L+NP F +G + P+G+L+ G G
Sbjct: 43 KELNKEVVPEKNVKTFKDVKGCDDAKQELEEVVEYLKNPAKFTRLGGKLPKGILLTGPPG 222
Query: 724 TGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFD 783
TGKT LA AIA EA VP + E M+VG A VR LFQ A+ AP I+F+++ D
Sbjct: 223 TGKTLLAKAIAGEAGVPFFYRAGSEFEE-MYVGVGARRVRSLFQAAKKKAPCIIFIDEID 399
Query: 784 LFAGVRGKFI-HTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRM 842
R ++ HT+ H QLLVE+DGFE+ +G++++A T +D AL RPGR
Sbjct: 400 AVGSTRKQWEGHTKKTLH-----QLLVEMDGFEQNEGIIVIAATNLPDILDPALTRPGRF 564
Query: 843 DRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
DR + P R+ IL D L + +D K +A T
Sbjct: 565 DRHIVVPNPDLRGRQEIL---ELYLQDKPLADDIDIKSIARGT 684
>TC206154 homologue to UP|FTSH_MEDSA (Q9BAE0) Cell division protein ftsH
homolog, chloroplast precursor , partial (88%)
Length = 2059
Score = 87.8 bits (216), Expect(2) = 3e-30
Identities = 46/114 (40%), Positives = 69/114 (60%)
Frame = +2
Query: 670 KRVKKPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSL 729
+ V + + + + D K E+ EVV FL+NP + +GA+ P+G L+VG GTGKT L
Sbjct: 491 QEVPETGVSFADVAGADQAKLELQEVVDFLKNPDKYTALGAKIPKGCLLVGPPGTGKTLL 670
Query: 730 AMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFD 783
A A+A EA VP A + ++VG AS VR+LF+ A+ AP I+F+++ D
Sbjct: 671 ARAVAGEAGVPFFSCAASEF-VELFVGVGASRVRDLFEKAKGKAPCIVFIDEID 829
Score = 63.9 bits (154), Expect(2) = 3e-30
Identities = 36/89 (40%), Positives = 50/89 (55%)
Frame = +1
Query: 797 NQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAER 856
N + E INQLL E+DGF GV+++A T +D AL RPGR DR + RP A R
Sbjct: 871 NDEREQTINQLLTEMDGFSGNSGVIVLAATNRPDVLDSALLRPGRFDRQVTVDRPDVAGR 1050
Query: 857 ENILYSAAKETMDDQLVEYVDWKKVAEKT 885
IL ++ L + VD++K+A +T
Sbjct: 1051VKILQVHSR---GKALAKDVDFEKIARRT 1128
>TC206116 UP|CC48_SOYBN (P54774) Cell division cycle protein 48 homolog
(Valosin containing protein homolog) (VCP), complete
Length = 2583
Score = 124 bits (310), Expect = 3e-28
Identities = 69/215 (32%), Positives = 115/215 (53%), Gaps = 1/215 (0%)
Frame = +1
Query: 672 VKKPPIPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAPRGVLIVGERGTGKTSLA 730
V+ P + + ++++K E+ E V + +++P F++ G +GVL G G GKT LA
Sbjct: 1438 VEVPNVSWEDIGGLENVKRELQETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLA 1617
Query: 731 MAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRG 790
AIA E + + +K +L MW G+S +NVRE+F AR AP +LF ++ D A RG
Sbjct: 1618 KAIANECQANFISVKGPELLT-MWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRG 1794
Query: 791 KFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQR 850
+ + +NQLL E+DG + V ++ T ID AL RPGR+D++ ++
Sbjct: 1795 SSVGDAGGAADRVLNQLLTEMDGMSAKKTVFIIGATNRPDIIDPALLRPGRLDQLIYIPL 1974
Query: 851 PTQAERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
P + R I + +++ + + VD + +A T
Sbjct: 1975 PDEDSRHQIFKACLRKS---PIAKNVDLRALARHT 2070
Score = 97.4 bits (241), Expect = 3e-20
Identities = 70/264 (26%), Positives = 133/264 (49%), Gaps = 18/264 (6%)
Frame = +1
Query: 667 EHMKRVKKPPIPLNNFSSIDSMKEEISEVVAF----LQNPRAFQEMGARAPRGVLIVGER 722
E +KR + + + + ++++++++ L++P+ F+ +G + P+G+L+ G
Sbjct: 595 EPLKREDEERLDEVGYDDVGGVRKQMAQIRELVELPLRHPQLFKSIGVKPPKGILLYGPP 774
Query: 723 GTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDF 782
G+GKT +A A+A E I ++ + + G+S SN+R+ F+ A AP I+F+++
Sbjct: 775 GSGKTLIARAVANETGAFFFCINGPEIMSKL-AGESESNLRKAFEEAEKNAPSIIFIDEI 951
Query: 783 DLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRM 842
D A R K T + ++QLL +DG + + V+++ T ID AL+R GR
Sbjct: 952 DSIAPKREK---THGEVERRIVSQLLTLMDGLKSRAHVIVIGATNRPNSIDPALRRFGRF 1122
Query: 843 DRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDWKKVAEKT--------------ALL 888
DR + P + R +L T + +L + VD +++A+ T A L
Sbjct: 1123DREIDIGVPDEVGRLEVL---RIHTKNMKLSDDVDLERIAKDTHGYVGADLAALCTEAAL 1293
Query: 889 RPIELKLVPIALEGSAFRSKVLDT 912
+ I K+ I LE ++VL++
Sbjct: 1294QCIREKMDVIDLEDETIDAEVLNS 1365
>TC225592 homologue to GB|AAP78936.1|32306505|BT009668 At5g19990 {Arabidopsis
thaliana;} , partial (94%)
Length = 1612
Score = 116 bits (291), Expect = 5e-26
Identities = 79/241 (32%), Positives = 124/241 (50%), Gaps = 3/241 (1%)
Frame = +3
Query: 659 VDPIKTAFEHMKRVKKPPIPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAPRGVL 717
VDP+ MK K P + +D +EI EV+ +++P F+ +G P+GVL
Sbjct: 525 VDPLVNL---MKVEKVPDSTYDMIGGLDQQIKEIKEVIELPIKHPELFESLGIAQPKGVL 695
Query: 718 IVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVIL 777
+ G GTGKT LA A+A + + +L ++G+ + VRELF AR+ AP I+
Sbjct: 696 LYGPPGTGKTLLARAVAHHTDCTFIRVSGSEL-VQKYIGEGSRMVRELFVMAREHAPSII 872
Query: 778 FVEDFDLFAGVRGKFIHTENQDHEA--FINQLLVELDGFEKQDGVVLMATTRNLKQIDEA 835
F+++ D R + + N D E + +LL +LDGFE + + ++ T + +D+A
Sbjct: 873 FMDEIDSIGSARMES-GSGNGDSEVQRTMLELLNQLDGFEASNKIKVLMATNRIDILDQA 1049
Query: 836 LQRPGRMDRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDWKKVAEKTALLRPIELKL 895
L RPGR+DR P + R +IL ++ L+ +D KK+AEK ELK
Sbjct: 1050LLRPGRIDRKIEFPNPNEESRLDILKIHSRRM---NLMRGIDLKKIAEKMNGASGAELKA 1220
Query: 896 V 896
V
Sbjct: 1221V 1223
>TC214213 homologue to UP|PRSA_BRACM (O23894) 26S protease regulatory subunit
6A homolog (TAT-binding protein homolog 1) (TBP-1),
partial (64%)
Length = 1086
Score = 108 bits (269), Expect = 2e-23
Identities = 66/223 (29%), Positives = 119/223 (52%), Gaps = 4/223 (1%)
Frame = +1
Query: 678 PLNNFSSIDSMKEEISEVVAFLQNPRA----FQEMGARAPRGVLIVGERGTGKTSLAMAI 733
P +++ I ++++I E+V + P FQ++G R P+GVL+ G GTGKT +A A
Sbjct: 34 PTEDYNDIGGLEKQIQELVEAIVLPMTHKERFQKLGVRPPKGVLLYGPPGTGKTLMARAC 213
Query: 734 AAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFI 793
AA+ +++ QL M++G A VR+ FQ A++ +P I+F+++ D R
Sbjct: 214 AAQTNATFLKLAGPQL-VQMFIGDGAKLVRDAFQLAKEKSPCIIFIDEIDAIGTKRFDSE 390
Query: 794 HTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQ 853
+ +++ + + +LL +LDGF D + ++A T +D AL R GR+DR P++
Sbjct: 391 VSGDREVQRTMLELLNQLDGFSSDDRIKVIAATNRADILDPALMRSGRLDRKIEFPHPSE 570
Query: 854 AERENILYSAAKETMDDQLVEYVDWKKVAEKTALLRPIELKLV 896
R IL +++ + V+++++A T +LK V
Sbjct: 571 EARARILQIHSRKM---NVHPDVNFEELARSTDDFNGAQLKAV 690
>TC225683 homologue to UP|Q9SZD4 (Q9SZD4) 26S proteasome subunit 4-like
protein (26S proteasome subunit AtRPT2a), partial (97%)
Length = 1513
Score = 105 bits (261), Expect = 2e-22
Identities = 69/225 (30%), Positives = 117/225 (51%), Gaps = 4/225 (1%)
Frame = +2
Query: 656 REGVDPIKTAFEHMKRVKKPPIPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAPR 714
++ VDP+ + MK K P + +D+ +EI E V L +P ++++G + P+
Sbjct: 509 QDEVDPMVSV---MKVEKAPLESYADIGGLDAQIQEIKEAVELPLTHPELYEDIGIKPPK 679
Query: 715 GVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAP 774
GV++ GE GTGKT LA A+A + + +L ++G VRELF+ A DL+P
Sbjct: 680 GVILYGEPGTGKTLLAKAVANSTSATFLRVVGSEL-IQKYLGDGPKLVRELFRVADDLSP 856
Query: 775 VILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDE 834
I+F+++ D R ++ + + +LL +LDGF+ + V ++ T ++ +D
Sbjct: 857 SIVFIDEIDAVGTKRYDAHSGGEREIQRTMLELLNQLDGFDSRGDVKVILATNRIESLDP 1036
Query: 835 ALQRPGRMDRIFHLQRPTQAERENI--LYSAAKETMDD-QLVEYV 876
AL RPGR+DR P R I ++++ DD L E+V
Sbjct: 1037ALLRPGRIDRKIEFPLPDIKTRRRIFQIHTSRMTLADDVNLEEFV 1171
>TC225681 homologue to UP|Q9SZD4 (Q9SZD4) 26S proteasome subunit 4-like
protein (26S proteasome subunit AtRPT2a), partial (93%)
Length = 1433
Score = 105 bits (261), Expect = 2e-22
Identities = 69/225 (30%), Positives = 117/225 (51%), Gaps = 4/225 (1%)
Frame = +1
Query: 656 REGVDPIKTAFEHMKRVKKPPIPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAPR 714
++ VDP+ + MK K P + +D+ +EI E V L +P ++++G + P+
Sbjct: 406 QDEVDPMVSV---MKVEKAPLESYADIGGLDAQIQEIKEAVELPLTHPELYEDIGIKPPK 576
Query: 715 GVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAP 774
GV++ GE GTGKT LA A+A + + +L ++G VRELF+ A DL+P
Sbjct: 577 GVILYGEPGTGKTLLAKAVANSTSATFLRVVGSEL-IQKYLGDGPKLVRELFRVADDLSP 753
Query: 775 VILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDE 834
I+F+++ D R ++ + + +LL +LDGF+ + V ++ T ++ +D
Sbjct: 754 SIVFIDEIDAVGTKRYDAHSGGEREIQRTMLELLNQLDGFDSRGDVKVILATNRIESLDP 933
Query: 835 ALQRPGRMDRIFHLQRPTQAERENI--LYSAAKETMDD-QLVEYV 876
AL RPGR+DR P R I ++++ DD L E+V
Sbjct: 934 ALLRPGRIDRKIEFPLPDIKTRRRIFQIHTSRMTLADDVNLEEFV 1068
>TC205434 homologue to UP|PRS6_ARATH (Q9SEI4) 26S protease regulatory subunit
6B homolog (26S proteasome AAA-ATPase subunit RPT3)
(Regulatory particle triple-A ATPase subunit 3), partial
(96%)
Length = 1573
Score = 102 bits (255), Expect = 8e-22
Identities = 61/212 (28%), Positives = 110/212 (51%), Gaps = 4/212 (1%)
Frame = +1
Query: 669 MKRVKKPPIPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAPRGVLIVGERGTGKT 727
+ + +KP + N+ D K+EI E V L + ++++G PRGVL+ G GTGKT
Sbjct: 544 LSQSEKPDVTYNDIGGCDIQKQEIREAVELPLTHHELYKQIGIDPPRGVLLYGPPGTGKT 723
Query: 728 SLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAG 787
LA A+A + + + ++G+ VR++F+ A++ AP I+F+++ D A
Sbjct: 724 MLAKAVANHTTAAFIRVVGSEF-VQKYLGEGPRMVRDVFRLAKENAPAIIFIDEVDAIAT 900
Query: 788 VRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFH 847
R +++ + + +LL ++DGF++ V ++ T +D AL RPGR+DR
Sbjct: 901 ARFDAQTGADREVQRILMELLNQMDGFDQTVNVKVIMATNRADTLDPALLRPGRLDRKIE 1080
Query: 848 LQRPTQAERENIL-YSAAKETMDDQ--LVEYV 876
P + ++ + AK + D+ L +YV
Sbjct: 1081FPLPDRRQKRLVFQVCTAKMNLSDEVDLEDYV 1176
>TC228291 homologue to UP|Q9FXT8 (Q9FXT8) 26S proteasome regulatory particle
triple-A ATPase subunit4, partial (62%)
Length = 1079
Score = 102 bits (254), Expect = 1e-21
Identities = 61/185 (32%), Positives = 100/185 (53%)
Frame = +3
Query: 699 LQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQS 758
L NP F +G + P+GVL+ G GTGKT LA AIA+ + +++ + + ++G+S
Sbjct: 36 LMNPELFIRVGIKPPKGVLLYGPPGTGKTLLARAIASNIEANFLKVVSSAI-IDKYIGES 212
Query: 759 ASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQD 818
A +RE+F ARD P I+F+++ D G R + +++ + + +LL +LDGF++
Sbjct: 213 ARLIREMFGYARDHQPCIIFMDEIDAIGGRRFSEGTSADREIQRTLMELLNQLDGFDQLG 392
Query: 819 GVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDW 878
V ++ T +D AL RPGR+DR + P + R IL A ++Y
Sbjct: 393 KVKMIMATNRPDVLDPALLRPGRLDRKIEIPLPNEQSRMEILKIHAAGIAKHGEIDYEAV 572
Query: 879 KKVAE 883
K+AE
Sbjct: 573 VKLAE 587
>BE661034 similar to SP|Q9ZPR1|C48B Cell division control protein 48 homolog
B (AtCDC48b). [Mouse-ear cress] {Arabidopsis thaliana},
partial (38%)
Length = 882
Score = 102 bits (254), Expect = 1e-21
Identities = 66/193 (34%), Positives = 101/193 (52%), Gaps = 2/193 (1%)
Frame = +1
Query: 704 AFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVR 763
AF MG RG+L+ G G KT+LA A A A+ + +L + M+VG+ + +R
Sbjct: 13 AFSXMGISPVRGILLHGPPGCSKTTLAKAAAHAAQASFFSLSGAELYS-MYVGEGEALLR 189
Query: 764 ELFQTARDLAPVILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLM 823
+ FQ AR AP I+F ++ D+ A RG E ++ LL E+DG E+ G++++
Sbjct: 190 KTFQRARLAAPSIIFFDEADVVAAKRGDSSSNSATVGERLLSTLLTEIDGLEEAKGILVL 369
Query: 824 ATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENIL--YSAAKETMDDQLVEYVDWKKV 881
A T ID AL RPGR D + ++ P R IL ++ +T +D VD +++
Sbjct: 370 AATNRPYAIDAALMRPGRFDLVLYVPPPDLEARHEILCVHTRKMKTGND-----VDLRRI 534
Query: 882 AEKTALLRPIELK 894
AE T L EL+
Sbjct: 535 AEDTELFTGAELE 573
>TC227714 similar to UP|C48C_ARATH (Q9SS94) Cell division control protein 48
homolog C (AtCDC48c), partial (22%)
Length = 918
Score = 95.9 bits (237), Expect = 9e-20
Identities = 60/160 (37%), Positives = 87/160 (53%)
Frame = +1
Query: 715 GVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAP 774
G L+ G G GKT +A A+A EA + IK +L +VG+S VR +F AR AP
Sbjct: 10 GFLLYGPPGCGKTLIAKAVANEAGATFIHIKGPEL-LNKYVGESELAVRTMFSRARTCAP 186
Query: 775 VILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDE 834
ILF ++ D RGK E +NQLLVELDG E++ GV ++ T + +D
Sbjct: 187 CILFFDEIDALTTKRGK---EGGWVVERLLNQLLVELDGAEQRKGVFVIGATNRPEVMDR 357
Query: 835 ALQRPGRMDRIFHLQRPTQAERENILYSAAKETMDDQLVE 874
A+ RPGR ++ ++ P+ ER IL + A++ D V+
Sbjct: 358 AVLRPGRFGKLLYVPLPSPDERVLILKALARKKAVDASVD 477
>AW201375 homologue to GP|17065470|gb cell division protein FtsH-like protein
{Arabidopsis thaliana}, partial (20%)
Length = 385
Score = 93.2 bits (230), Expect = 6e-19
Identities = 51/131 (38%), Positives = 76/131 (57%)
Frame = +3
Query: 726 KTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLF 785
KT LA A+A EA VP + A + + VG+ A+ +R+LF AR AP I+F+++ D
Sbjct: 3 KTLLARAVAGEAGVPFFTVSASEF-VELVVGRGAARIRDLFNAARKFAPSIIFIDELDAV 179
Query: 786 AGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRI 845
G RG+ N + + +NQLL E+DGFE + VV++A T + +D AL RPGR R
Sbjct: 180 GGKRGRSF---NDERDQTLNQLLTEMDGFESEMRVVVIAATNRPEALDPALCRPGRFSRK 350
Query: 846 FHLQRPTQAER 856
++ P + R
Sbjct: 351 VYVGEPDEEGR 383
>TC215551 homologue to UP|PRS7_SPIOL (Q41365) 26S protease regulatory subunit
7 (26S proteasome subunit 7) (26S proteasome AAA-ATPase
subunit RPT1) (Regulatory particle triple-A ATPase
subunit 1), complete
Length = 1561
Score = 92.8 bits (229), Expect = 8e-19
Identities = 66/223 (29%), Positives = 104/223 (46%), Gaps = 1/223 (0%)
Frame = +3
Query: 638 EQMAYYFDERKGRMRDRRREGVDPIKTAFEHMKRVKKPPIPLNNFSSIDSMKEEISEVVA 697
E M D K +++ +DP T M +KP + N+ E++ EVV
Sbjct: 441 EGMRVGVDRNKYQIQIPLPPKIDPSVTM---MTVEEKPDVTYNDVGGCKEQIEKMREVVE 611
Query: 698 F-LQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVG 756
+ +P F ++G P+GVL G GTGKT LA A+A + + +L +VG
Sbjct: 612 LPMLHPEKFVKLGIDPPKGVLCYGPPGTGKTLLARAVANRTDACFIRVIGSEL-VQKYVG 788
Query: 757 QSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEK 816
+ A VRELFQ AR I+F ++ D G R + + + + +++ +LDGF+
Sbjct: 789 EGARMVRELFQMARSKKACIVFFDEVDAIGGARFDDGVGGDNEVQRTMLEIVNQLDGFDA 968
Query: 817 QDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENI 859
+ + ++ T +D AL RPGR+DR P R I
Sbjct: 969 RGNIKVLMATNRPDTLDPALLRPGRLDRKVEFGLPDLESRTQI 1097
>TC215552 UP|PRS7_ARATH (Q9SSB5) 26S protease regulatory subunit 7 (26S
proteasome subunit 7) (26S proteasome AAA-ATPase subunit
RPT1a) (Regulatory particle triple-A ATPase subunit 1a),
partial (73%)
Length = 1082
Score = 92.8 bits (229), Expect = 8e-19
Identities = 66/223 (29%), Positives = 104/223 (46%), Gaps = 1/223 (0%)
Frame = +1
Query: 638 EQMAYYFDERKGRMRDRRREGVDPIKTAFEHMKRVKKPPIPLNNFSSIDSMKEEISEVVA 697
E M D K +++ +DP T M +KP + N+ E++ EVV
Sbjct: 34 EGMRVGVDRNKYQIQIPLPPKIDPSVTM---MTVEEKPDVTYNDVGGCKEQIEKMREVVE 204
Query: 698 F-LQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVG 756
+ +P F ++G P+GVL G GTGKT LA A+A + + +L +VG
Sbjct: 205 LPMLHPEKFVKLGIDPPKGVLCYGPPGTGKTLLARAVANRTDACFIRVIGSEL-VQKYVG 381
Query: 757 QSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEK 816
+ A VRELFQ AR I+F ++ D G R + + + + +++ +LDGF+
Sbjct: 382 EGARMVRELFQMARSKKACIVFFDEVDAIGGARFDDGVGGDNEVQRTMLEIVNQLDGFDA 561
Query: 817 QDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENI 859
+ + ++ T +D AL RPGR+DR P R I
Sbjct: 562 RGNIKVLMATNRPDTLDPALLRPGRLDRKVEFGLPDLESRTQI 690
>TC205252 homologue to UP|Q9ZP50 (Q9ZP50) FtsH-like protein Pftf precursor,
partial (61%)
Length = 1482
Score = 58.5 bits (140), Expect(2) = 1e-18
Identities = 33/68 (48%), Positives = 42/68 (61%)
Frame = +1
Query: 723 GTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDF 782
GTGKT LA AIA EA VP I + M+VG AS VR+LF+ A++ AP I+FV++
Sbjct: 1 GTGKTLLAKAIAGEAGVPFFSISGSEF-VEMFVGVGASRVRDLFKKAKENAPCIVFVDEI 177
Query: 783 DLFAGVRG 790
D RG
Sbjct: 178 DAVGRQRG 201
Score = 53.9 bits (128), Expect(2) = 1e-18
Identities = 27/64 (42%), Positives = 36/64 (56%)
Frame = +3
Query: 797 NQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAER 856
N + E +NQLL E+DGFE G++++A T +D AL RPGR DR + P R
Sbjct: 222 NDEREQTLNQLLTEMDGFEGNTGIIVVAATNRADILDSALLRPGRFDRQVTVDVPDIRGR 401
Query: 857 ENIL 860
IL
Sbjct: 402 TEIL 413
>TC206370 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, partial (38%)
Length = 1502
Score = 90.5 bits (223), Expect = 4e-18
Identities = 72/232 (31%), Positives = 113/232 (48%), Gaps = 13/232 (5%)
Frame = +2
Query: 667 EHMKRVKKPPIPLN-------NFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAPRGVLI 718
E KR++ IP N + ++D +KE + E+V L+ P F+ + RG+L+
Sbjct: 2 EFEKRIRPEVIPANEIGVTFADIGALDEIKESLQELVMLPLRRPDLFKGGLLKPCRGILL 181
Query: 719 VGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILF 778
G GTGKT LA AIA EA + + + + W G+ NVR LF A +AP I+F
Sbjct: 182 FGPPGTGKTMLAKAIANEAGASFINVSMSTITS-KWFGEDEKNVRALFTLAAKVAPTIIF 358
Query: 779 VEDFDLFAGVRGKFIHTENQDHEA---FINQLLVELDGF--EKQDGVVLMATTRNLKQID 833
V++ D G R T +HEA N+ + DG + ++++A T +D
Sbjct: 359 VDEVDSMLGQR-----TRVGEHEAMRKIKNEFMTHWDGLLTGPNEQILVLAATNRPFDLD 523
Query: 834 EALQRPGRMDRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
EA+ R R +R + P+ RE IL K + + E +D+K++A T
Sbjct: 524 EAIIR--RFERRILVGLPSVENREMIL----KTLLAKEKHENLDFKELATMT 661
>TC229555 similar to GB|AAM62497.1|21553404|AY085265 26S proteasome
regulatory particle chain RPT6-like protein {Arabidopsis
thaliana;} , partial (85%)
Length = 1155
Score = 89.4 bits (220), Expect = 9e-18
Identities = 63/204 (30%), Positives = 103/204 (49%), Gaps = 8/204 (3%)
Frame = +2
Query: 677 IPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAP-RGVLIVGERGTGKTSLAMAIA 734
+ N+ ++++K+ + E+V L+ P F P +GVL+ G GTGKT LA AIA
Sbjct: 233 VEFNSIGGLETIKQALFELVILPLKRPDLFSHGKLLGPQKGVLLYGPPGTGKTMLAKAIA 412
Query: 735 AEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIH 794
E++ + ++ L W G + V +F A L P I+F+++ D F G R
Sbjct: 413 KESRAVFINVRISNL-MSKWFGDAQKLVAAVFSLAYKLQPAIIFIDEVDSFLGKR----- 574
Query: 795 TENQDHEAFIN---QLLVELDGF--EKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQ 849
DHEA +N + + DGF ++ V+++A T ++DEA+ R R+ + F +
Sbjct: 575 -RGTDHEAMLNMKTEFMALWDGFTTDQNAQVMVLAATNRPSELDEAILR--RLPQAFEIG 745
Query: 850 RPTQAERENILYSAAK-ETMDDQL 872
P Q ER IL K E ++D +
Sbjct: 746 IPDQRERAEILKVVLKGERVEDNI 817
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.332 0.143 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 51,017,089
Number of Sequences: 63676
Number of extensions: 727588
Number of successful extensions: 5827
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 5634
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5786
length of query: 1184
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1076
effective length of database: 5,762,624
effective search space: 6200583424
effective search space used: 6200583424
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC135461.15


