

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135461.13 + phase: 0
(560 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
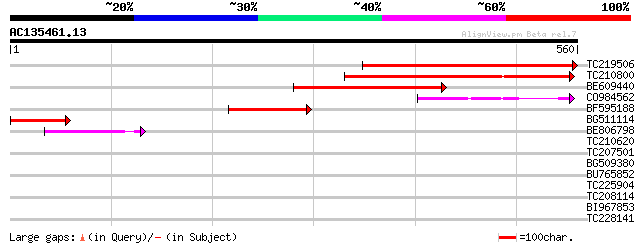
Score E
Sequences producing significant alignments: (bits) Value
TC219506 similar to UP|Q9M8Z1 (Q9M8Z1) T6K12.3 protein (AT3g0435... 404 e-113
TC210800 similar to UP|Q9M8Z1 (Q9M8Z1) T6K12.3 protein (AT3g0435... 347 1e-95
BE609440 weakly similar to GP|20197184|gb expressed protein {Ara... 187 1e-47
CO984562 120 2e-27
BF595188 similar to GP|15450691|gb| AT3g04350/T6K12_3 {Arabidops... 112 4e-25
BG511114 104 9e-23
BE806798 similar to GP|20197184|gb| expressed protein {Arabidops... 76 5e-14
TC210620 40 0.003
TC207501 similar to GB|AAN38679.1|23507751|BT000842 At4g34190/F2... 30 2.2
BG509380 29 4.9
BU765852 29 6.3
TC225904 similar to UP|O48937 (O48937) NADPH cytochrome P450 red... 29 6.3
TC208114 similar to UP|DME_ARATH (Q8LK56) Transcriptional activa... 28 8.3
BI967853 28 8.3
TC228141 similar to GB|AAL77730.1|18700240|AY078029 AT3g54810/F2... 28 8.3
>TC219506 similar to UP|Q9M8Z1 (Q9M8Z1) T6K12.3 protein (AT3g04350/T6K12_3),
partial (36%)
Length = 995
Score = 404 bits (1037), Expect = e-113
Identities = 186/212 (87%), Positives = 201/212 (94%)
Frame = +3
Query: 349 LGGAFTDIAMWVFCPFNGPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFF 408
LGGAFTDI MWVFCPFNGPATLKV+LMNIEM+KIGEHVGDWEHFTLR+SNFTGELWSV+F
Sbjct: 3 LGGAFTDIVMWVFCPFNGPATLKVALMNIEMSKIGEHVGDWEHFTLRISNFTGELWSVYF 182
Query: 409 SEHSGGKWVNAFDLEFIKENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNF 468
S+HSGG W++AFDLEF K NKPIVYSS+ GHAS+PH GTYLQGSSKLGIGVRNDAA S F
Sbjct: 183 SQHSGGGWIHAFDLEFNKGNKPIVYSSKDGHASFPHPGTYLQGSSKLGIGVRNDAAPSKF 362
Query: 469 ILDSSFRYKIVAAEYLGDGVITEPCWLQYMREWGPTIVYDSRSEIEKIIDMLPIFVRFSV 528
I+DSS +Y+IVAAEYLGDGVI EPCWLQYMREWGPTIVYDSRSEIEKII++LP+FVRFSV
Sbjct: 363 IVDSSIKYQIVAAEYLGDGVIAEPCWLQYMREWGPTIVYDSRSEIEKIINLLPLFVRFSV 542
Query: 529 ENLFELFPTELSGEEGPTGPKEKDNWLGDEYC 560
ENLFELFPTEL GEEGPTGPKEKDNWLGDEYC
Sbjct: 543 ENLFELFPTELYGEEGPTGPKEKDNWLGDEYC 638
>TC210800 similar to UP|Q9M8Z1 (Q9M8Z1) T6K12.3 protein (AT3g04350/T6K12_3),
partial (35%)
Length = 986
Score = 347 bits (889), Expect = 1e-95
Identities = 155/228 (67%), Positives = 197/228 (85%)
Frame = +1
Query: 331 LKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNGPATLKVSLMNIEMNKIGEHVGDWE 390
++ G+++SA+LYVHVKPALGG FTDIAMWVFCPFNGP+TLK+ + + +K+GEHVGDWE
Sbjct: 10 IQAGDLKSAKLYVHVKPALGGTFTDIAMWVFCPFNGPSTLKIGITSRAFSKVGEHVGDWE 189
Query: 391 HFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFIKENKPIVYSSRHGHASYPHAGTYLQ 450
HFTLR+ NF+GELWS++FS+HSGGKWV+A++LE+I NK +VYSS++GHASYPH GTYLQ
Sbjct: 190 HFTLRICNFSGELWSIYFSQHSGGKWVDAYELEYIDGNKAVVYSSKNGHASYPHPGTYLQ 369
Query: 451 GSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPCWLQYMREWGPTIVYDSR 510
GSSKLGIG+RNDA +SN +DSS +Y++VAAEYLGD V+ EP WLQ+MREWGP IVYDS+
Sbjct: 370 GSSKLGIGIRNDATRSNLYVDSSVQYELVAAEYLGD-VVREPQWLQFMREWGPKIVYDSK 546
Query: 511 SEIEKIIDMLPIFVRFSVENLFELFPTELSGEEGPTGPKEKDNWLGDE 558
+E++KII+ LP +R S +L + P EL GEEGPTGPKEK+NW+GDE
Sbjct: 547 TELDKIINALPGGLRNSFVSLIKKLPVELYGEEGPTGPKEKNNWIGDE 690
>BE609440 weakly similar to GP|20197184|gb expressed protein {Arabidopsis
thaliana}, partial (26%)
Length = 461
Score = 187 bits (475), Expect = 1e-47
Identities = 88/152 (57%), Positives = 107/152 (69%), Gaps = 1/152 (0%)
Frame = +1
Query: 281 WFFKNGAILYTAGN-AKGKAIDYHGTNLPGGGYNDGAFWIDLPTDEDARSNLKKGNIESA 339
WFF NGA+LY G +K +I +G NLP DGA+W+DLP D + +KKG+++SA
Sbjct: 1 WFFSNGALLYKKGQESKPVSISPNGANLPQDPNIDGAYWVDLPADSTNKERVKKGDLKSA 180
Query: 340 ELYVHVKPALGGAFTDIAMWVFCPFNGPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNF 399
YVHVKP LGG FTDIAMWVF PFNGPA KV + + + KIGEHVGDWEH TLRVSNF
Sbjct: 181 ISYVHVKPMLGGTFTDIAMWVFYPFNGPARAKVEFLTVNLGKIGEHVGDWEHVTLRVSNF 360
Query: 400 TGELWSVFFSEHSGGKWVNAFDLEFIKENKPI 431
GEL V+FS+HS G W ++ LEF NKP+
Sbjct: 361 NGELKHVYFSQHSKGVWFDSSQLEFQSGNKPL 456
>CO984562
Length = 623
Score = 120 bits (301), Expect = 2e-27
Identities = 65/156 (41%), Positives = 84/156 (53%)
Frame = -1
Query: 403 LWSVFFSEHSGGKWVNAFDLEFIKENKPIVYSSRHGHASYPHAGTYLQGSSKLGIGVRND 462
LW V+FS+HS G+W +A +LEF N+P+ YSS HGHA +P G +QG G+GVRND
Sbjct: 623 LWKVYFSQHSKGQWEDASELEFQNGNRPVAYSSLHGHALFPKPGLVMQGMR--GLGVRND 450
Query: 463 AAKSNFILDSSFRYKIVAAEYLGDGVITEPCWLQYMREWGPTIVYDSRSEIEKIIDMLPI 522
AAKS+ ++D + ++IVAAEYLG I EP WL + WGP
Sbjct: 449 AAKSDAVMDMATWFEIVAAEYLG-SQIREPPWLNFCMNWGP------------------- 330
Query: 523 FVRFSVENLFELFPTELSGEEGPTGPKEKDNWLGDE 558
+EGP GPK+KD W GDE
Sbjct: 329 -------------------KEGPKGPKQKDMWKGDE 279
>BF595188 similar to GP|15450691|gb| AT3g04350/T6K12_3 {Arabidopsis
thaliana}, partial (10%)
Length = 249
Score = 112 bits (280), Expect = 4e-25
Identities = 51/82 (62%), Positives = 65/82 (79%)
Frame = +2
Query: 217 VSVGTFFCGTYFDSEQVVDVVCLKNLDSLLHAMPNLNQIHALIEHYGPTVYFHPDEKYMP 276
VSVGTFFC + +++ + VVCLKNL+ +L AMP L+ IHALIEHYGPTV+FHP+E Y+P
Sbjct: 2 VSVGTFFCTSCWNNGDELSVVCLKNLNPVLPAMPRLH*IHALIEHYGPTVFFHPEEVYLP 181
Query: 277 SSVSWFFKNGAILYTAGNAKGK 298
SSV WFF NGA+LY G + G+
Sbjct: 182 SSVDWFFNNGALLYRKGVSTGE 247
>BG511114
Length = 357
Score = 104 bits (260), Expect = 9e-23
Identities = 47/60 (78%), Positives = 49/60 (81%)
Frame = +2
Query: 1 MFGFECFCWDSFPEFFDSDPLPFSLPSPLPQWPQGGGFAGGRISLGKIEVTKVNKFEKVW 60
MFG + CWDS PEF D DPLPFSLPSPLPQWPQGG A GRI LG+IEV KVNKFEKVW
Sbjct: 176 MFGCKSLCWDSVPEFTDPDPLPFSLPSPLPQWPQGGSXACGRICLGEIEVIKVNKFEKVW 355
>BE806798 similar to GP|20197184|gb| expressed protein {Arabidopsis
thaliana}, partial (17%)
Length = 412
Score = 75.9 bits (185), Expect = 5e-14
Identities = 40/101 (39%), Positives = 57/101 (55%), Gaps = 1/101 (0%)
Frame = +1
Query: 35 GGGFAGGRISLGK-IEVTKVNKFEKVWRCTNSNGKALGFTFYRPLEIPDGFSCLGYYCHS 93
GGGFA G I LG + V++++ F KVW LG TF+ P + +GF LG YC
Sbjct: 94 GGGFATGIIDLGGGLLVSQISTFNKVWTTYEGGPNNLGATFFEPTGLSEGFFMLGCYCQP 273
Query: 94 NDQPLRGHVLVARETTSKSQADCSESESPALKKPLNYSLIW 134
N++PL G VLV ++ +S S AL +P++Y L+W
Sbjct: 274 NNKPLHGCVLVGKDNSSTSNG--------ALAEPVDYKLVW 372
>TC210620
Length = 698
Score = 40.0 bits (92), Expect = 0.003
Identities = 20/65 (30%), Positives = 30/65 (45%), Gaps = 2/65 (3%)
Frame = +1
Query: 127 PLNYSLIW--CMDSHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVC 184
P+ Y L+W C + + + W P P G+ A G V EP+ + V C+ L E
Sbjct: 28 PMGYDLVWRNCPEDYVTPLSIWHPRAPDGFVAPGCVAIAGYLEPEPDLVYCIAESLVEET 207
Query: 185 ETSDL 189
E +L
Sbjct: 208 EFEEL 222
>TC207501 similar to GB|AAN38679.1|23507751|BT000842 At4g34190/F28A23_50
{Arabidopsis thaliana;} , partial (52%)
Length = 724
Score = 30.4 bits (67), Expect = 2.2
Identities = 11/28 (39%), Positives = 18/28 (64%)
Frame = +2
Query: 62 CTNSNGKALGFTFYRPLEIPDGFSCLGY 89
C+NS G L +FY P+ + + +CLG+
Sbjct: 452 CSNSTGGCLDSSFYLPISLKELITCLGW 535
>BG509380
Length = 238
Score = 29.3 bits (64), Expect = 4.9
Identities = 12/38 (31%), Positives = 20/38 (52%)
Frame = -3
Query: 35 GGGFAGGRISLGKIEVTKVNKFEKVWRCTNSNGKALGF 72
GGGF+ G ++L +E T + F + K++GF
Sbjct: 131 GGGFSNGAVALANVEATGIVPFTAIVLFEGEEFKSIGF 18
>BU765852
Length = 427
Score = 28.9 bits (63), Expect = 6.3
Identities = 12/40 (30%), Positives = 21/40 (52%)
Frame = +2
Query: 435 SRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSF 474
+ H H + H L KLG+ +RN AA+ + +L + +
Sbjct: 164 THHHHLEFSHYNNPLHSVDKLGMRMRNSAARVSLLLGNFY 283
>TC225904 similar to UP|O48937 (O48937) NADPH cytochrome P450 reductase ,
partial (68%)
Length = 1704
Score = 28.9 bits (63), Expect = 6.3
Identities = 16/53 (30%), Positives = 24/53 (45%), Gaps = 1/53 (1%)
Frame = +1
Query: 252 LNQIHALIEHYGPTVYFHPDEKYMPSSVSW-FFKNGAILYTAGNAKGKAIDYH 303
L+++ GPT + + +S W GA +Y G+AKG A D H
Sbjct: 1156 LDELILAFSREGPTKEYVQHKMMEKASEIWSMISQGAYIYVCGDAKGMARDVH 1314
>TC208114 similar to UP|DME_ARATH (Q8LK56) Transcriptional activator DEMETER
(DNA glycosylase-related protein DME), partial (6%)
Length = 704
Score = 28.5 bits (62), Expect = 8.3
Identities = 13/35 (37%), Positives = 15/35 (42%), Gaps = 7/35 (20%)
Frame = -3
Query: 6 CFCWDSFPEFFDSDPLPFSLP-------SPLPQWP 33
C+CW S P D +P LP S L WP
Sbjct: 423 CYCWQSIPAVLDLVSIPVILPLNSTPFLSSLLSWP 319
>BI967853
Length = 666
Score = 28.5 bits (62), Expect = 8.3
Identities = 13/42 (30%), Positives = 21/42 (49%)
Frame = +2
Query: 208 CDRGMLARGVSVGTFFCGTYFDSEQVVDVVCLKNLDSLLHAM 249
C ++ RG+ + T F + ++CL NL LLHA+
Sbjct: 380 CSYSLIGRGILIXX*LAHTRFGRKLPTFLLCLNNLYLLLHAL 505
>TC228141 similar to GB|AAL77730.1|18700240|AY078029 AT3g54810/F28P10_210
{Arabidopsis thaliana;} , partial (23%)
Length = 1143
Score = 28.5 bits (62), Expect = 8.3
Identities = 12/36 (33%), Positives = 19/36 (52%)
Frame = +1
Query: 435 SRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFIL 470
+ H H + H L KLG+ +RN AA+ + +L
Sbjct: 229 THHHHLEFSHYNNPLHSVDKLGMRMRNSAARVSLLL 336
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.139 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,281,213
Number of Sequences: 63676
Number of extensions: 471698
Number of successful extensions: 2413
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 2383
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2409
length of query: 560
length of database: 12,639,632
effective HSP length: 102
effective length of query: 458
effective length of database: 6,144,680
effective search space: 2814263440
effective search space used: 2814263440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC135461.13


